For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTVAIGPGAGPSSTRPSSQPSDLHSGLRAVTECTGSAVVVHVGGDIDASNEVAWQRLVSKSAAIAIAPG PFVIDIRDLDFMGSCAYAVLAQESVRCRRRGVNMRLVSNQPIVARTIAACGLRRLIPLYAMVETALAPPP SAH
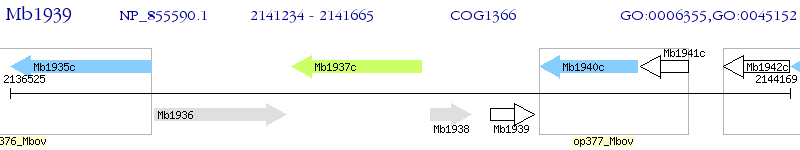
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1939 | - | - | 100% (143) | hypothetical protein Mb1939 |
| M. bovis AF2122 / 97 | Mb2671 | - | 8e-34 | 52.00% (125) | hypothetical protein Mb2671 |
| M. bovis AF2122 / 97 | Mb1400c | - | 9e-28 | 47.15% (123) | hypothetical protein Mb1400c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0203 | - | 7e-08 | 26.32% (133) | anti-sigma-factor antagonist |
| M. tuberculosis H37Rv | Rv1904 | - | 9e-78 | 99.30% (143) | hypothetical protein Rv1904 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2800 | - | 3e-62 | 79.72% (143) | hypothetical protein MMAR_2800 |
| M. avium 104 | MAV_3518 | - | 3e-33 | 50.81% (124) | anti-anti-sigma factor |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2925 | - | 2e-62 | 79.72% (143) | hypothetical protein MUL_2925 |
| M. vanbaalenii PYR-1 | Mvan_2530 | - | 1e-08 | 34.26% (108) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1939|M.bovis_AF2122/97 ----MRTVAIGPGAGPSSTRPSSQPSDLH-SGLRAVTECTGSAVVVHVGG
Rv1904|M.tuberculosis_H37Rv ----MRTVAIGPGAGPSSTRPSSQPSDLH-SGLRAVTECTGSAVVVHVGG
MMAR_2800|M.marinum_M -MRRVNTVAMGSSPSSSSTRLSSPAGDPH-TGLRAVTECTGSAVVVHVGG
MUL_2925|M.ulcerans_Agy99 ----MNTVAMGSSPSSSSTRLSSPAGDPH-TGLRAVTECTGSAVVVHVGG
MAV_3518|M.avium_104 ------MEPMSATHLTLSTKLVYELGDPN-STLRATTARSGSAVLIYAGG
Mvan_2530|M.vanbaalenii_PYR-1 ------MVAVGTKTIYRPLTAVTDEQERCGHATFAVRRCSAHRMAIAVIG
Mflv_0203|M.gilvum_PYR-GCK MAISEALPPSSTQRVFRPDPTSFELREEHHRATFSACELPPTTVLVTIHG
. .. . : :. . : : *
Mb1939|M.bovis_AF2122/97 DIDASNEVAWQRLVSKSAAIAIAPGPFVIDIRDLDFMGSCAYAVLAQESV
Rv1904|M.tuberculosis_H37Rv DIDASNEVAWQRLVSKSAAIAIAPGPFVIDIRDLDFMGSCAYAVLAQESV
MMAR_2800|M.marinum_M DVDASNEVVWQRLVNRSAAIAIAPGPFVIDIRDLDFIGSCAYAVLAQESV
MUL_2925|M.ulcerans_Agy99 DVDASNEVVWQRLVNRSAAIAIAPGPFVIDIRDLDFIGSCAYAVLAQESV
MAV_3518|M.avium_104 EIDACNEDTWRHLVSEAAGVVATPGPFVVDVTGLEFMGCCAFAVLTDEAK
Mvan_2530|M.vanbaalenii_PYR-1 EIDAVNGRALGRYVERHTRISRQ---LVLDLRAVDFFGSQGFTALYYISV
Mflv_0203|M.gilvum_PYR-GCK EIDATNNMPLAHYIEKRIGDSRK---LIVDLQTVEFFAASGFAALFNVNV
::** * : :.. :::*: ::*:.. .::.*
Mb1939|M.bovis_AF2122/97 RCRRRGVNMRLVSNQPIVARTIAACGLRRLIPLYAMVETALAPPPSAH--
Rv1904|M.tuberculosis_H37Rv RCRRRGVNMRLVSNQPIVARTIAACGLRRLIPLYATVETALAPPPSAH--
MMAR_2800|M.marinum_M RCRRRGVNLRLVSNQPIVARTIAACGLRRLLPMYSSVEAALSPVPNGH--
MUL_2925|M.ulcerans_Agy99 RCRRRGVNLRLVSNQPIVARTIAACGLRRLLPMYSSVEAALSPVPNGH--
MAV_3518|M.avium_104 RCRQRGIDLRLVSCEPIVGRIIDACGLSDILPIYPTVDSALSGADRW---
Mvan_2530|M.vanbaalenii_PYR-1 HCARSDVDWVIVG-SPAVRRLLAICDPDGELPLTDALSSALARLDRLAHG
Mflv_0203|M.gilvum_PYR-GCK VCHRRGVRWQLLA-GPHVRRLLRVCDPQRELPVTGSSAKYTRTRPRDRK-
* : .: ::. * * * : *. :*:
Mb1939|M.bovis_AF2122/97 ---------------
Rv1904|M.tuberculosis_H37Rv ---------------
MMAR_2800|M.marinum_M ---------------
MUL_2925|M.ulcerans_Agy99 ---------------
MAV_3518|M.avium_104 ---------------
Mvan_2530|M.vanbaalenii_PYR-1 GQHLVWTAKSGWHAR
Mflv_0203|M.gilvum_PYR-GCK ------LLVGGYH--