For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAPDSITAVVTDHDGVTVLSIGGEIDLVTAPALEEAIGAVVADSPTALVIDLSGVEFLGSVGLKILAATY EKLGTAAEFGVVAQGPATRRPIHLTGLDKTFPLYPALDDALTGVRDGKLNR*
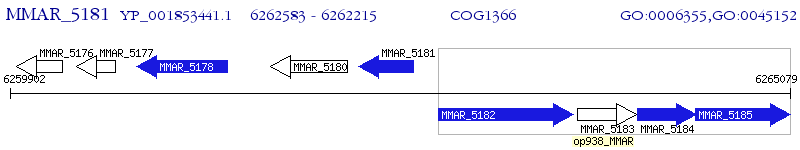
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5181 | - | - | 100% (122) | anti-anti-sigma regulatory factor |
| M. marinum M | MMAR_0214 | - | 6e-21 | 44.44% (99) | hypothetical protein MMAR_0214 |
| M. marinum M | MMAR_4567 | - | 2e-07 | 27.05% (122) | hypothetical protein MMAR_4567 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3712c | - | 6e-29 | 74.07% (81) | hypothetical protein Mb3712c |
| M. gilvum PYR-GCK | Mflv_2021 | - | 2e-21 | 46.94% (98) | anti-sigma-factor antagonist |
| M. tuberculosis H37Rv | Rv3687c | rsfB | 2e-47 | 76.23% (122) | anti-anti-sigma factor RSFB (anti-sigma factor antagonist) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_0435 | - | 3e-50 | 85.71% (112) | anti-anti-sigma factor |
| M. smegmatis MC2 155 | MSMEG_6127 | - | 5e-21 | 43.00% (100) | anti-anti-sigma factor |
| M. thermoresistible (build 8) | TH_1832 | - | 1e-28 | 55.05% (109) | ANTI-ANTI-SIGMA FACTOR RSFB (ANTI-SIGMA FACTOR ANTAGONIST) |
| M. ulcerans Agy99 | MUL_4427 | - | 2e-07 | 27.05% (122) | hypothetical protein MUL_4427 |
| M. vanbaalenii PYR-1 | Mvan_3003 | - | 3e-21 | 41.96% (112) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5181|M.marinum_M -----------MSAPDSITAVVTDH--DGVTVLSIGGEIDLVTAPALEEA
MAV_0435|M.avium_104 --------------------MVEDH--DGVSVVSVSGEIDLVTAPALEQA
Mb3712c|M.bovis_AF2122/97 --------------------------------------------------
Rv3687c|M.tuberculosis_H37Rv -----------MSAPDSITVTVADH--NGVAVLSIGGEIDLITAAALEEA
TH_1832|M.thermoresistible__bu ---------------------VEHR--DDSAVLRVQGVVDLATGDALQQA
MSMEG_6127|M.smegmatis_MC2_155 MTSQ-----------DPANCTVEERRVGDITVVAVTGTVDMLTAPKLEDA
Mvan_3003|M.vanbaalenii_PYR-1 MDEQAAEGTNVGGAASSSTCVVTERWVERTAVVAVSGVVDMLTSPQLESA
Mflv_2021|M.gilvum_PYR-GCK ------------------MQQFTEREVDGVVIVAAVGAIDMLTAPQLQEA
MUL_4427|M.ulcerans_Agy99 ---------MNATAKSPNALAIDTRSEDSLVVLSVEGAIDSTNCAALRDA
MMAR_5181|M.marinum_M IGAVVADSPTALVIDLSGVEFLGSVGLKILAATYEKLGT--AAEFGVVAQ
MAV_0435|M.avium_104 IGAVVADSPPALVIDLSAVEFLGSVGLKILAATYEKLGK--ETGFGVVAR
Mb3712c|M.bovis_AF2122/97 ----MADNPTALVIDLSAVEFLGSVGLKILAATSEKIGQ--SVKFGVVAR
Rv3687c|M.tuberculosis_H37Rv IGEVVADNPTALVIDLSAVEFLGSVGLKILAATSEKIGQ--SVKFGVVAR
TH_1832|M.thermoresistible__bu IDDLIATRPSALVVDLSAVDFLASVGLQILVATHERLSP--SARFAVVAD
MSMEG_6127|M.smegmatis_MC2_155 IGSAAKSEPSAVVVDLSAVDFLASAGMGVLVAAHGELAP--AVRLVVVAD
Mvan_3003|M.vanbaalenii_PYR-1 IDTALSQQPAAVVIDFTDVEFLASAGMGVLVAAHDKAGS--DVKISVVAE
Mflv_2021|M.gilvum_PYR-GCK VAAAARRRPSGLVVDLTSVDFLGSAGMQVLMETRKGLDG--DTRFAVVAD
MUL_4427|M.ulcerans_Agy99 IIKATLDEPSAVVVNVSALQVPDEASWSIFVSARWQVDTPQHVPILLVCA
*..:*::.: ::. ... :: : . : :*.
MMAR_5181|M.marinum_M GPATRRPIHLTGLDKTFPLYPALDDALTGVRDGKLNR-------------
MAV_0435|M.avium_104 GPATRRPIHLTGLDKTFPLYPTLDDALTAVRDGKLNG-------------
Mb3712c|M.bovis_AF2122/97 GSVTRRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------------
Rv3687c|M.tuberculosis_H37Rv GSVTRRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------------
TH_1832|M.thermoresistible__bu GPATSRPIQLTRLDEVIALYPTLDEALSGVRTGERAE-------------
MSMEG_6127|M.smegmatis_MC2_155 GPATSRPLKLVGIADVVDLFATLDEALSSLKT------------------
Mvan_3003|M.vanbaalenii_PYR-1 GPATSRPLKLVGIADIVALYPNLEEALAARDT------------------
Mflv_2021|M.gilvum_PYR-GCK GPATSRPLKITGITDYVDLYPSLDVALADLAG------------------
MUL_4427|M.ulcerans_Agy99 SRAGRELITRTGVTRFMPVYPTEKRAIKAVGRLARRKVRYAQVQLPANLG
. . . : : . ::. . *:
MMAR_5181|M.marinum_M --------------------------------------------------
MAV_0435|M.avium_104 --------------------------------------------------
Mb3712c|M.bovis_AF2122/97 --------------------------------------------------
Rv3687c|M.tuberculosis_H37Rv --------------------------------------------------
TH_1832|M.thermoresistible__bu --------------------------------------------------
MSMEG_6127|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3003|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2021|M.gilvum_PYR-GCK --------------------------------------------------
MUL_4427|M.ulcerans_Agy99 SLRESRKLVREWLTSWSKPGLIPVALVIVNVFVENVLEHTGSDPVIKVQC
MMAR_5181|M.marinum_M --------------------------------------------------
MAV_0435|M.avium_104 --------------------------------------------------
Mb3712c|M.bovis_AF2122/97 --------------------------------------------------
Rv3687c|M.tuberculosis_H37Rv --------------------------------------------------
TH_1832|M.thermoresistible__bu --------------------------------------------------
MSMEG_6127|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_3003|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2021|M.gilvum_PYR-GCK --------------------------------------------------
MUL_4427|M.ulcerans_Agy99 DGTAATIAVSDGSNAPAIRLQTPPKGIDVSGLAIVEALCRAWGSTPTATG
MMAR_5181|M.marinum_M -------------
MAV_0435|M.avium_104 -------------
Mb3712c|M.bovis_AF2122/97 -------------
Rv3687c|M.tuberculosis_H37Rv -------------
TH_1832|M.thermoresistible__bu -------------
MSMEG_6127|M.smegmatis_MC2_155 -------------
Mvan_3003|M.vanbaalenii_PYR-1 -------------
Mflv_2021|M.gilvum_PYR-GCK -------------
MUL_4427|M.ulcerans_Agy99 KTVWAIIGPENQL