For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SVDIAEPFTPHPMLSIRLVRELGDPHSTLRATTDFRSAAVLIHAGGEVDAANEHTWRQLIAETAASAPSP GLFIVDVSGLDFMGCCAFEVLAEQAGRCRARGVELRIASAAHTRVTRIVAACGLSDALPVYRSVDAALAL PAPSD*
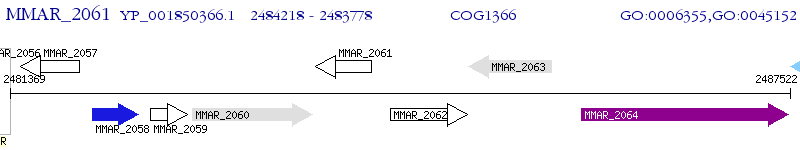
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2061 | - | - | 100% (146) | hypothetical protein MMAR_2061 |
| M. marinum M | MMAR_2800 | - | 2e-31 | 50.00% (130) | hypothetical protein MMAR_2800 |
| M. marinum M | MMAR_5181 | - | 6e-07 | 31.67% (120) | anti-anti-sigma regulatory factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2671 | - | 8e-42 | 63.85% (130) | hypothetical protein Mb2671 |
| M. gilvum PYR-GCK | Mflv_4424 | - | 7e-07 | 30.91% (110) | anti-sigma-factor antagonist |
| M. tuberculosis H37Rv | Rv2638 | - | 9e-42 | 63.85% (130) | hypothetical protein Rv2638 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3518 | - | 7e-40 | 59.69% (129) | anti-anti-sigma factor |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2508 | - | 3e-05 | 28.44% (109) | PUTATIVE putative anti-sigma-factor antagonist |
| M. ulcerans Agy99 | MUL_3282 | - | 9e-79 | 97.26% (146) | hypothetical protein MUL_3282 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2061|M.marinum_M MSVDIAEPFT-PHPMLSIRLVRELGDPHSTLRATTDFRSAAVLIHAGGEV
MUL_3282|M.ulcerans_Agy99 MSVDIAEPFT-PHPMLSIRLLRELGDPHSTLRAATDFRSTAVLIHAGGEV
Mb2671|M.bovis_AF2122/97 MGLITTEPRSSPHP-LSPRLVHELGDPHSTLRATTDGSGAALLIHAGGEI
Rv2638|M.tuberculosis_H37Rv MGLITTEPRSSPHP-LSPRLVHELGDPHSTLRATTDGSGAALLIHAGGEI
MAV_3518|M.avium_104 -----MEPMSATHLTLSTKLVYELGDPNSTLRATTARSGSAVLIYAGGEI
Mflv_4424|M.gilvum_PYR-GCK -----------------------MAERVGLLEIGQEVHDVAVVVRAGGEV
TH_2508|M.thermoresistible__bu ----MILPVPRPSP-----GGGEAVTCHAADFATRWVNSATVVIRVDGEL
. . :::: ..**:
MMAR_2061|M.marinum_M DAANEHTWRQLIAETAASAPSP--GLFIVDVSGLDFMGCCAFEVLAEQAG
MUL_3282|M.ulcerans_Agy99 DAANEHTWRQLIAETAASAPSP--GLFVVDVSGLDFMGCCAFEVLAEQAG
Mb2671|M.bovis_AF2122/97 DGRNEHLWRQLVTEAAAGVTAP--GPLIVDVTGLDFMGCCAFAALADEAQ
Rv2638|M.tuberculosis_H37Rv DGRNEHLWRQLVTEAAAGVTAP--GPLIVDVTGLDFMGCCAFAALADEAQ
MAV_3518|M.avium_104 DACNEDTWRHLVSEAAGVVATP--GPFVVDVTGLEFMGCCAFAVLTDEAK
Mflv_4424|M.gilvum_PYR-GCK DSGTVETLAAALDSAVAAAAAHPTRTVVVDLDGITYFGSAGLNALLTCRE
TH_2508|M.thermoresistible__bu DAANSPQLVDYALRHAGRIKYL-----VADLTGVEFFGIAGFSAAHDING
*. . .. :.*: *: ::* ..: .
MMAR_2061|M.marinum_M RCRARGVELRIASAAHTRVTRIVAACGLSDALPVYRSVDAALA--LPAPS
MUL_3282|M.ulcerans_Agy99 RCRARGVELRIASAAHTRVTRIVAACGLSDALPVYRSVDAALA--LPAPS
Mb2671|M.bovis_AF2122/97 RCRCRGIDLRLVS-HQPIVARIAEAGGLSRVLPIYPTVDTALGKGTAGPA
Rv2638|M.tuberculosis_H37Rv RCRCRGIDLRLVS-HQPIVARIAEAGGLSRVLPIYPTVDTALGKGTAGPA
MAV_3518|M.avium_104 RCRQRGIDLRLVS-CEPIVGRIIDACGLSDILPIYPTVDSAL----SGAD
Mflv_4424|M.gilvum_PYR-GCK EGEGRGVVVRLVA-TTAEVTRPIEVTRLARILRPYPSVGEAVAAPEDPPE
TH_2508|M.thermoresistible__bu RCAHAKLTWTLVP--SPAVVRLLQICDPDGTLPLNTTVPDSVWDNGGSGG
. : :.. . * * * :* ::
MMAR_2061|M.marinum_M D--------
MUL_3282|M.ulcerans_Agy99 D--------
Mb2671|M.bovis_AF2122/97 RC-------
Rv2638|M.tuberculosis_H37Rv RC-------
MAV_3518|M.avium_104 RW-------
Mflv_4424|M.gilvum_PYR-GCK DPR------
TH_2508|M.thermoresistible__bu LFQLIPEAG