For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRNENVAGLLAERASEAGWTDQPAYYAPDVVTHGQIHDGAARLGAVLANRGLCRGDRVLLCMPDSPELVQ VLLACLARGILAFLANPELHRDDHAFQERDTQAALVITSGPLCDRFAPSTVVDAADLFSEAARVGPADYE ILGGDAAAYATYTSGTTGPPKAAIHRHCDVFAFVEAMCRNALRLTPADIGLSSARMYFAYGLGNSVWFPL ATGSSAVVNPLPVGAEVAATLSARFEPSVLYGVPNFFARVVDACSADSFRSVRCVVSAGEALEVGLAERL TEFFGGIPILDGVGSTEVGQTFVSNTVDEWRPGSLGKVLPPYQIRVVAPDGAAAGPGVEGDLWVRGPSIA ESYWNWPEPLLTDEGWLDTRDRVCIDDDGWVTYACRADDTEIVGAVNINPREIERLIVEEDAVAEVAVVG VKEATGASTLQAFLVPASAEGIDGSVMRDIHRRLLTRLSAFKVPHRFAVVERLPRTANGKLLRSALRGQT PAKPIWELASAEHRSGAPGQLDDQSASALVSGSREVSLKERLAALQQERHRLVLDAVCGETAKMLGEPDP RSVNRDLAFSELGFDSQMTVELCHRLAAATGLRVPETVGWDYGSISGLAQYLEAELSGADRRVTPQSARS GARALPLIEAQLNKVEELTAAIADGEKPRVAERLRALLGTITEGQEHWGQRIAAASTPDEIFQLIDSEFG ES
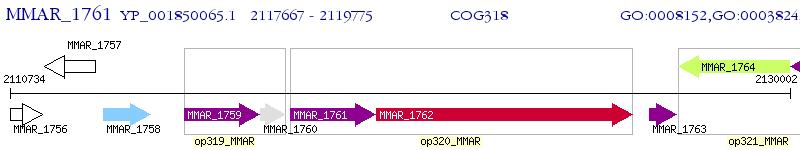
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1761 | fadD22 | - | 100% (702) | fatty-acid-CoA ligase FadD22 |
| M. marinum M | MMAR_0101 | - | 0.0 | 65.05% (641) | polyketide synthase PKS |
| M. marinum M | MMAR_1606 | - | 2e-43 | 28.51% (470) | fatty-acid-CoA ligase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2972c | fadD22 | 0.0 | 74.61% (705) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_4616 | - | 2e-34 | 27.31% (476) | AMP-dependent synthetase and ligase |
| M. tuberculosis H37Rv | Rv2948c | fadD22 | 0.0 | 74.61% (705) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_00134 | fadD22 | 0.0 | 72.84% (707) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_4569c | - | 9e-37 | 28.16% (515) | fatty-acid-CoA ligase FadD |
| M. avium 104 | MAV_5018 | - | 3e-35 | 27.98% (486) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_5633 | - | 2e-34 | 27.50% (509) | feruloyl-CoA synthetase |
| M. thermoresistible (build 8) | TH_3626 | - | 4e-40 | 29.09% (464) | PUTATIVE putative fatty-acid-CoA ligase |
| M. ulcerans Agy99 | MUL_2004 | fadD22 | 0.0 | 99.15% (702) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_0131 | - | 9e-37 | 28.35% (515) | acyl-CoA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_5018|M.avium_104 -MLSLTAQLAH-------------HPTQANEQPYLSRRQNWVNQLERHAL
Mvan_0131|M.vanbaalenii_PYR-1 -MFDQISSFRNPSMEQKTLTEAIQQPGFATEQPYLARRQNWTNQLARHAL
MAB_4569c|M.abscessus_ATCC_199 -MTDTIDATVQ---------------SAGLAQPYRARRQNWCNQLARHAL
Mflv_4616|M.gilvum_PYR-GCK -MDRGIGQWVT-----------------------------------KRAF
TH_3626|M.thermoresistible__bu -MLSQSVHLVS--------------------------------LPDQRAA
MSMEG_5633|M.smegmatis_MC2_155 --MNLFGLLDQ-----------------------------------TAAR
MMAR_1761|M.marinum_M MRNENVAGLLA----------------------------------ERASE
MUL_2004|M.ulcerans_Agy99 MRNENVAGLLA----------------------------------ERASE
Mb2972c|M.bovis_AF2122/97 MRNGNLAGLLA----------------------------------EQASE
Rv2948c|M.tuberculosis_H37Rv MRNGNLAGLLA----------------------------------EQASE
MLBr_00134|M.leprae_Br4923 MRSENLAALLA----------------------------------RQAAE
:
MAV_5018|M.avium_104 MQPNATALRFLGKGLTWGELHGRVRALADALSRRGVGFGDRVMVLMLNRP
Mvan_0131|M.vanbaalenii_PYR-1 MQPDQPALRFLGNTTTWGELDRRVSALAGALHKRGVGFGDRVLILMLNRT
MAB_4569c|M.abscessus_ATCC_199 MQPDATAIRYLGHSISWAELHQRVQALADALSRRGVGFGDRVLILMLNRP
Mflv_4616|M.gilvum_PYR-GCK LNGQRTALVQGDTSFTYSDFDRRTNQVASSLLRLGVRKGDRVAILLVNSV
TH_3626|M.thermoresistible__bu SAPAAPCLADDTVALSNAEFRHAVDSAAAVLRGHGVGAGDVVAVMLPNTV
MSMEG_5633|M.smegmatis_MC2_155 HGDRGAVFCGERELYTWTQLRDRAARLGSTLTEP----GTRVAVASENRP
MMAR_1761|M.marinum_M AGWTDQPAYYAPDVVTHGQIHDGAARLGAVLANRGLCRGDRVLLCMPDSP
MUL_2004|M.ulcerans_Agy99 AGWTDQPAYYAPDVVTHGQIHDGAARLGAVLANRGLCRGDRVLLCMPDSP
Mb2972c|M.bovis_AF2122/97 AGWYDRPAFYAADVVTHGQIHDGAARLGEVLRNRGLSSGDRVLLCLPDSP
Rv2948c|M.tuberculosis_H37Rv AGWYDRPAFYAADVVTHGQIHDGAARLGEVLRNRGLSSGDRVLLCLPDSP
MLBr_00134|M.leprae_Br4923 AGWYDKPAYFAPDVVTHGQIHDGAVRLGEVLRNRGLSAGDRVLLCLPDSP
: :: . . * * * : :
MAV_5018|M.avium_104 EFMESVLAINMLGAIAVPLNFRLTAAEIAFLVQDCQARVVITEAVLAPVA
Mvan_0131|M.vanbaalenii_PYR-1 EFIEAVLATNKLGAIAVPVNFRMTPAEIAFLVSDCEAKAIITEAVLANVA
MAB_4569c|M.abscessus_ATCC_199 EYVESWLAINELGAIAIPVNFRMTPPEVAFLVENSGAKVAITETVLAPVA
Mflv_4616|M.gilvum_PYR-GCK EFLEVLLGCAKIGAITIPINVRLAGPEIGYILADSGADVFVFHAPLAAAA
TH_3626|M.thermoresistible__bu DLVVTLFAAWRLGAAVTPLNPALSVDESAYQIADAGAKVLVAGGNLPAPA
MSMEG_5633|M.smegmatis_MC2_155 EIIEIMFGVWAAECVYVPINYKLHPREMADILADSGAALVFASPKIAPGL
MMAR_1761|M.marinum_M ELVQVLLACLARGILAFLANPELHRDDHAFQERDTQAALVITSGPLCDRF
MUL_2004|M.ulcerans_Agy99 ELVQVLLACLARGILAFLANPELHRDDQAFQERDTQAALVITSGPLCDRF
Mb2972c|M.bovis_AF2122/97 DLVQLLLACLARGVMAFLANPELHRDDHALAARNTEPALVVTSDALRDRF
Rv2948c|M.tuberculosis_H37Rv DLVQLLLACLARGVMAFLANPELHRDDHALAARNTEPALVVTSDALRDRF
MLBr_00134|M.leprae_Br4923 DLVQLLLACLARGIMAFLANPELHRDDYAFPERDTAAALVITNGSLRDRF
: : :. * : : . : . . :
MAV_5018|M.avium_104 T-GVRDIESLLDTVVVAGGSSDDAVLGYDDLIDETGAAHQPVDIPNDAAA
Mvan_0131|M.vanbaalenii_PYR-1 T-AVRDIDPTLATVISAGGSGDD-VLDYDDVLAEN-APCPVVDIPNDAPA
MAB_4569c|M.abscessus_ATCC_199 A-AVRQQAPALETVIVAGSASENGVLGYEEVIAEPGEPHAEVDLPGDTPA
Mflv_4616|M.gilvum_PYR-GCK VSALTEPGVRVRHTLRAGGAPADGEIPYTDLLSDGAPAPLDSDVEGRDPA
TH_3626|M.thermoresistible__bu TPVVRPADLKTGHGPVEVAPPAD-----------------------DRLA
MSMEG_5633|M.smegmatis_MC2_155 FEVTDVPVEVVGTDAYTQRFESE--------------PVPAPLTDPAELA
MMAR_1761|M.marinum_M APSTVVDAADLFSEAARVGPADY------------------EILGGDAAA
MUL_2004|M.ulcerans_Agy99 APSTVVDAADLFSEAARAGPADY------------------EILGGDAAA
Mb2972c|M.bovis_AF2122/97 QPSRVAEAAELMSEAARVAPGGY------------------EPMGGDALA
Rv2948c|M.tuberculosis_H37Rv QPSRVAEAAELMSEAARVAPGGY------------------EPMGGDALA
MLBr_00134|M.leprae_Br4923 QSSNVVEPAELLSDATRVEPSDY------------------EPVSGDAYA
*
MAV_5018|M.avium_104 LIMYTSGTTGRPKGAVLTHTNLTGQTMTGLYTNGADINNDVGFIGVPFFH
Mvan_0131|M.vanbaalenii_PYR-1 LIMYTSGTTGRPKGAVLTHTNLTGQVMTLLFTNGADINHDIGFIGVPFFH
MAB_4569c|M.abscessus_ATCC_199 LIMYTSGTTGRPKGAVLTHLNLSGQAMTYLLSTTVDLNSDVGFIGVPMFH
Mflv_4616|M.gilvum_PYR-GCK FIMYTSGTTGRPKGAILTHDNLRWNAINVLGAEQGLTGSDVTVAVAPMFH
TH_3626|M.thermoresistible__bu LLIYTSGTTGRPKGVMLDHANIAAMCASIIEAN-ELTSADSSLLILPLFH
MSMEG_5633|M.smegmatis_MC2_155 WLFYTSGTTGKSKGAMLSHRNLMAMTVAHLADFDAPDEFCSLVHGAPMSH
MMAR_1761|M.marinum_M YATYTSGTTGPPKAAIHRHCDVFAFVEAMCRNALRLTPADIGLSSARMYF
MUL_2004|M.ulcerans_Agy99 YATYTSGTTGPPKAAIHRHCDVFAFVEAMCCNALRLTPADIGLSSARMYF
Mb2972c|M.bovis_AF2122/97 YATYTSGTTGPPKAAIHRHADPLTFVDAMCRKALRLTPEDTGLCSARMYF
Rv2948c|M.tuberculosis_H37Rv YATYTSGTTGPPKAAIHRHADPLTFVDAMCRKALRLTPEDTGLCSARMYF
MLBr_00134|M.leprae_Br4923 FATYTSGTTGKPKAAIHRHADPFTFVDAMCRKALRLTPQDIGLCSARMYF
******* .*..: * : . : .
MAV_5018|M.avium_104 IAGIG-NMLTGLLLGIPTVIYPLGAFEPGQLLDVLAAEKVTGIFLVPAQW
Mvan_0131|M.vanbaalenii_PYR-1 IAGVG-SSVSGLLLGRPTVLYPVGAFDPNELLDVLEAEKVTGIFLVPAQW
MAB_4569c|M.abscessus_ATCC_199 IAGVG-NMITGLLLGLPTVIHPLGAFDPGALLDVLESEGVTGIFLVPAQW
Mflv_4616|M.gilvum_PYR-GCK IGGLGVHTLPLLYVGGTSVILPS--FDPVGTLKAMAESRATVQFMVPAMW
TH_3626|M.thermoresistible__bu VNGIVVGTLSPLIAGGHVTIAGR--FDPRTFFDRVRRSRPTYFSAVPTIY
MSMEG_5633|M.smegmatis_MC2_155 GSGLY--IPPYVLRGARQVIPESGAFEPDEFLDLCDHHPGCSAFLAPTMV
MMAR_1761|M.marinum_M AYGLGNSVWFPLATGSSAVVNPLP-VGAEVAATLSARFEPSVLYGVPNFF
MUL_2004|M.ulcerans_Agy99 AYGLGNSVWFPLATGSSAVVNPLP-VGAEVAATLSARFEPSVLYGVPNFF
Mb2972c|M.bovis_AF2122/97 AYGLGNSVWFPLATGGSAVINSAP-VTPEAAAILSARFGPSVLYGVPNFF
Rv2948c|M.tuberculosis_H37Rv AYGLGNSVWFPLATGGSAVINSAP-VTPEAAAILSARFGPSVLYGVPNFF
MLBr_00134|M.leprae_Br4923 AYGLGNSVWFPLATGGSAVISSVP-VSAESAAMLSTRFEPSVLYGVPSFF
*: : * .: . . .*
MAV_5018|M.avium_104 QAVCAEQRARPRDLKLRVISWG-AAPAPDALLREMSAMFPGTQILAAFGQ
Mvan_0131|M.vanbaalenii_PYR-1 QAVCAAQKARPRDLKLRFLSWG-AAPASDTLLREMAETFPGAQILAAFGQ
MAB_4569c|M.abscessus_ATCC_199 QAVCAAQQANPRKVRLKTLSWG-AAPATDVLLRTMSETFPDARILAAFGQ
Mflv_4616|M.gilvum_PYR-GCK SALTQVPDFDSYDLSALRLAMGGGAPMPLTVIDFMHQR--GVPFTEGFGM
TH_3626|M.thermoresistible__bu TKLLELPEDPTVDTSSLRFAACGAAPASAELLQRFEQR-YGVPIIEGYGL
MSMEG_5633|M.smegmatis_MC2_155 QRLIQTGRPRPANLRTVVYGGG---PMYVDSLKKALAA-YGQIFVQLYGQ
MMAR_1761|M.marinum_M ARVVDACSADSFRSVRCVVSAG--EALEVGLAERLTEFFGGIPILDGVGS
MUL_2004|M.ulcerans_Agy99 ARVVDACSADSFRSVRCVVSAG--EALEVGLAERLTEFFGGIPILDGVGS
Mb2972c|M.bovis_AF2122/97 ARVIDSCSPDSFRSLRCVVSAG--EALELGLAERLMEFFGGIPILDGIGS
Rv2948c|M.tuberculosis_H37Rv ARVIDSCSPDSFRSLRCVVSAG--EALELGLAERLMEFFGGIPILDGIGS
MLBr_00134|M.leprae_Br4923 ARVVGACSPDSFRSLRCVVTAG--EALEPALAERLVEFFGGIPILDGIGS
: . . . : *
MAV_5018|M.avium_104 TEMSPVTCMLLGEDAI----RKRGSVGKVIPTVAARVVDENMNDVPVGEV
Mvan_0131|M.vanbaalenii_PYR-1 TEMSPVTCMLLGDDAI----RKLGSVGKVIPTVSARIVDEDMNDVPVGQV
MAB_4569c|M.abscessus_ATCC_199 TEMSPVTCMLMGDDAI----RKMGSVGRVIPTVSARVVDDNMNDVPVGEV
Mflv_4616|M.gilvum_PYR-GCK TETAPMVSVLDAANIT----TRAGSIGRVAMHVDARIVDADDRDVPADTV
TH_3626|M.thermoresistible__bu SEG-TCVSTANPLRGP----RKPGTVGLPLPGQQVRIVDAEGRTLGANEI
MSMEG_5633|M.smegmatis_MC2_155 GEAPMTITGLRRDDHLDADDATLGSVGYPRSGVQVAVLRSDGTPADIGEI
MMAR_1761|M.marinum_M TEVGQTFVSNTVDEWR------PGSLGKVLPPYQIRVVAPDGAAAGPGVE
MUL_2004|M.ulcerans_Agy99 TEVGQTFVSNTVDEWR------PGSLGKVLPPYQIRVVAPDGAAAGPGGE
Mb2972c|M.bovis_AF2122/97 TEVGQTFVSNRVDEWR------LGTLGRVLPPYEIRVVAPDGTTAGPGVE
Rv2948c|M.tuberculosis_H37Rv TEVGQTFVSNRVDEWR------LGTLGRVLPPYEIRVVAPDGTTAGPGVE
MLBr_00134|M.leprae_Br4923 SEVGQTFVSNSVDDWR------VGTLGKVLPPYEIRVVAPDGATAGSGIE
* *::* :: : .
MAV_5018|M.avium_104 GEIVYRAPTLMSGYWNNPEATAEAFAGGWFHSGDLVRMDEDGYVWVVDRK
Mvan_0131|M.vanbaalenii_PYR-1 GEIVYRAPTLMAGYWNNPKATAEAFAGGWFHSGDLVRQDEEGYIWVVDRK
MAB_4569c|M.abscessus_ATCC_199 GEIVYRAPTLMQGYWNNPEATADAFAGGWFHSGDLVRQDEDGFVWVVDRK
Mflv_4616|M.gilvum_PYR-GCK GELLVRGPNVFVGYWMKPATTAEAFRGGWFHTGDLGRIDADGYITLVDRK
TH_3626|M.thermoresistible__bu GEVLIAGPTVMRGYLNRPEETAKTLVDGWLRTGDIGYLDDDGYLVLVDRA
MSMEG_5633|M.smegmatis_MC2_155 GEIVCRGDVVMSGYWNNPEATAATLKDGWLHTGDMGSFDARGHLTLRDRS
MMAR_1761|M.marinum_M GDLWVRGPSIAESYWNWPE--PLLTDEGWLDTRDRVCIDDDGWVTYACRA
MUL_2004|M.ulcerans_Agy99 GDLWVRGPSIAESYWNWPE--PLLTDEGWLDTRDRVCIDDDGWVTYACRA
Mb2972c|M.bovis_AF2122/97 GDLWVRGPAIAKGYWNRPD--SPVANEGWLDTRDRVCIDSDGWVTYRCRA
Rv2948c|M.tuberculosis_H37Rv GDLWVRGPAIAKGYWNRPD--SPVANEGWLDTRDRVCIDSDGWVTYRCRA
MLBr_00134|M.leprae_Br4923 GNLWVRGPSIAQSYWNRPD--SLLENGDWLNTRDRVRIDGDGWVTYGCRA
*:: . : .* * . .*: : * * * : *
MAV_5018|M.avium_104 KDMIISGGENIYCAEVENVLASHPDIVEVAVIGRAHEKWGEVPIAVAAVA
Mvan_0131|M.vanbaalenii_PYR-1 KDMIISGGENIYCAEVENVLAAHPEIVEVAVIGRAHEKWGEVPVAVAVVR
MAB_4569c|M.abscessus_ATCC_199 KDMIISGGENIYCAEVENALAEHEQIAEVAVIGRPHEKWGEVPVAVAAIH
Mflv_4616|M.gilvum_PYR-GCK KDMIISGGENVYPIEVEQVLFRHPGVLDAAVVGGPDDKWGERVVAVVVAD
TH_3626|M.thermoresistible__bu KDMIIRGGENIYPKEIEAVAHRLPEISEVAVVGRPHPLYGEVPVMYVAAA
MSMEG_5633|M.smegmatis_MC2_155 KDVVISGGSNIYPREVEEVLLEHPDVVEAGVVGAPDAEWGEIVVAFVVTN
MMAR_1761|M.marinum_M DDTEIVGAVNINPREIERLIVEEDAVAEVAVVGVKEATGASTLQAFLVPA
MUL_2004|M.ulcerans_Agy99 DDTEIVGAVNINPREIERLIVEEDAVAEVAVVGVKEATGASTLQAFLVPA
Mb2972c|M.bovis_AF2122/97 DDTEVIGGVNVDPREVERLIIEDEAVAEAAVVAVRESTGASTLQAFLVAT
Rv2948c|M.tuberculosis_H37Rv DDTEVIGGVNVDPREVERLIIEDEAVAEAAVVAVRESTGASTLQAFLVAT
MLBr_00134|M.leprae_Br4923 DDTEIVGGVNINPREVERLIIEADAVAEAAVVGVREFTGASTLQAFLVPA
.* : *. *: *:* : :..*:. . .. .
MAV_5018|M.avium_104 N-------DNLALEDLDEFLTERLARYKHPKALEIVDALPRNPAGKVLKT
Mvan_0131|M.vanbaalenii_PYR-1 SSLEATPGAGIELADLEQFLTERLARYKHPKALEIVDALPRNPAGKVLKT
MAB_4569c|M.abscessus_ATCC_199 G-------AALTLAELDGFLTARLARYKHPKDLVIVDALPRNPSGKVLKT
Mflv_4616|M.gilvum_PYR-GCK PAAE----QEPSADELIAWCRERLAHFKCPREVHLLPELPRNATGKLLKT
TH_3626|M.thermoresistible__bu PG------AVVDTDAVRRHLAVSLAKYKQPVAIRVLDVLPKNAVGKIDKP
MSMEG_5633|M.smegmatis_MC2_155 G-------TPVDAAALDAHLLERIARFKRPKRYEFIDALPKNSYGKVLKR
MMAR_1761|M.marinum_M SAEG---IDGSVMRDIHRRLLTRLSAFKVPHRFAVVERLPRTANGKLLRS
MUL_2004|M.ulcerans_Agy99 SAEG---IDGSVMRHIHRRLLTRLSAFKVPHRFAVVERLPRTANGKLLRS
Mb2972c|M.bovis_AF2122/97 SGAT---IDGSVMRDLHRGLLNRLSAFKVPHRFAVVDRLPRTPNGKLVRG
Rv2948c|M.tuberculosis_H37Rv SGAT---IDGSVMRDLHRGLLNRLSAFKVPHRFAVVDRLPRTPNGKLVRG
MLBr_00134|M.leprae_Br4923 VGAF---IDESVMRDVHRRLLTQLTAFKVPHRFAIIERLPRSTNGKLLRN
: :: :* * .: **:.. **: :
MAV_5018|M.avium_104 ELRIRYGGG-----------------------------------------
Mvan_0131|M.vanbaalenii_PYR-1 ELRIRYSAASFADAGESSTPPTISTAPQRN--------------------
MAB_4569c|M.abscessus_ATCC_199 ELRQRFGIAD----------------------------------------
Mflv_4616|M.gilvum_PYR-GCK ELRKRFTGVEGGVVQR----------------------------------
TH_3626|M.thermoresistible__bu AIRTLDATATQTA-------------------------------------
MSMEG_5633|M.smegmatis_MC2_155 ELRERVTG------------------------------------------
MMAR_1761|M.marinum_M ALRGQTPAKPIWELASAEHRSGAPGQLDDQSASA-----LVSGSREVSLK
MUL_2004|M.ulcerans_Agy99 ALRGQTPAKPIWELASAEHRSDAPGQLDDQSASA-----LVSGSREVSLK
Mb2972c|M.bovis_AF2122/97 ALRKQSPTKPIWELSLTEPGSGVRAQRDDLSASN--MTIAGGNDGGATLR
Rv2948c|M.tuberculosis_H37Rv ALRKQSPTKPIWELSLTEPGSGVRAQRDDLSASN--MTIAGGNDGGATLR
MLBr_00134|M.leprae_Br4923 VLRAQSPTKPIWELSLTESQSATKAQLDGRPASNAHAQAAVGHAAGATLK
:*
MAV_5018|M.avium_104 --------------------------------------------------
Mvan_0131|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4569c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4616|M.gilvum_PYR-GCK --------------------------------------------------
TH_3626|M.thermoresistible__bu --------------------------------------------------
MSMEG_5633|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1761|M.marinum_M ERLAALQQERHRLVLDAVCGETAKMLGEPDPRSVNRDLAFSELGFDSQMT
MUL_2004|M.ulcerans_Agy99 ERLAALQQERHRLVLDAVCGETAKMLGEPDPRSVNRDLAFSELGFDSQMT
Mb2972c|M.bovis_AF2122/97 ERLVALRQERQRLVVDAVCAEAAKMLGEPDPWSVDQDLAFSELGFDSQMT
Rv2948c|M.tuberculosis_H37Rv ERLVALRQERQRLVVDAVCAEAAKMLGEPDPWSVDQDLAFSELGFDSQMT
MLBr_00134|M.leprae_Br4923 QRLSALQQERERLVVEAVCAEAVKMLGESDPGLINRDLAFSDLGFDSQMT
MAV_5018|M.avium_104 --------------------------------------------------
Mvan_0131|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4569c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4616|M.gilvum_PYR-GCK --------------------------------------------------
TH_3626|M.thermoresistible__bu --------------------------------------------------
MSMEG_5633|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1761|M.marinum_M VELCHRLAAATGLRVPETVGWDYGSISGLAQYLEAELSGADRR-VTPQSA
MUL_2004|M.ulcerans_Agy99 VELCHRLAAATGLRVPETVGWDYGSISGLAQYLEAELSGADRR-VTPQSA
Mb2972c|M.bovis_AF2122/97 VTLCKRLAAVTGLRLPETVGWDYGSISGLAQYLEAELAGGHGRLKSAGPV
Rv2948c|M.tuberculosis_H37Rv VTLCKRLAAVTGLRLPETVGWDYGSISGLAQYLEAELAGGHGRLKSAGPV
MLBr_00134|M.leprae_Br4923 VTLCNRLAVVTGLRLPETVGWDYGSISGLSRYLEAELSGVRSRPETPLSA
MAV_5018|M.avium_104 --------------------------------------------------
Mvan_0131|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4569c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4616|M.gilvum_PYR-GCK --------------------------------------------------
TH_3626|M.thermoresistible__bu --------------------------------------------------
MSMEG_5633|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1761|M.marinum_M RSGARALPLIEAQLNKVEELTAAIADGEKPRVAERLRALLGTITEGQEHW
MUL_2004|M.ulcerans_Agy99 RSGARALPLIEAQLNKVEELTAAIADGEKPRVAERLRALLGTITEGQEHW
Mb2972c|M.bovis_AF2122/97 NSGATGLWAIEEQLNKVEELVAVIADGEKQRVADRLRALLGTIAGSEAGL
Rv2948c|M.tuberculosis_H37Rv NSGATGLWAIEEQLNKVEELVAVIADGEKQRVADRLRALLGTIAGSEAGL
MLBr_00134|M.leprae_Br4923 NSGAKGLSPIDEELKKVEEMVVAIGASEKQRVADRLRALLGIIVDGEAGL
MAV_5018|M.avium_104 ------------------------
Mvan_0131|M.vanbaalenii_PYR-1 ------------------------
MAB_4569c|M.abscessus_ATCC_199 ------------------------
Mflv_4616|M.gilvum_PYR-GCK ------------------------
TH_3626|M.thermoresistible__bu ------------------------
MSMEG_5633|M.smegmatis_MC2_155 ------------------------
MMAR_1761|M.marinum_M GQRIAAASTPDEIFQLIDSEFGES
MUL_2004|M.ulcerans_Agy99 GQRIAAASTPDEIFQLIDSEFGES
Mb2972c|M.bovis_AF2122/97 GKLIQAASTPDEIFQLIDSELGK-
Rv2948c|M.tuberculosis_H37Rv GKLIQAASTPDEIFQLIDSELGK-
MLBr_00134|M.leprae_Br4923 SKRIQAASTPDEIFQLIDSELCE-