For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSLTAQLAHHPTQANEQPYLSRRQNWVNQLERHALMQPNATALRFLGKGLTWGELHGRVRALADALSRRG VGFGDRVMVLMLNRPEFMESVLAINMLGAIAVPLNFRLTAAEIAFLVQDCQARVVITEAVLAPVATGVRD IESLLDTVVVAGGSSDDAVLGYDDLIDETGAAHQPVDIPNDAAALIMYTSGTTGRPKGAVLTHTNLTGQT MTGLYTNGADINNDVGFIGVPFFHIAGIGNMLTGLLLGIPTVIYPLGAFEPGQLLDVLAAEKVTGIFLVP AQWQAVCAEQRARPRDLKLRVISWGAAPAPDALLREMSAMFPGTQILAAFGQTEMSPVTCMLLGEDAIRK RGSVGKVIPTVAARVVDENMNDVPVGEVGEIVYRAPTLMSGYWNNPEATAEAFAGGWFHSGDLVRMDEDG YVWVVDRKKDMIISGGENIYCAEVENVLASHPDIVEVAVIGRAHEKWGEVPIAVAAVANDNLALEDLDEF LTERLARYKHPKALEIVDALPRNPAGKVLKTELRIRYGGG*
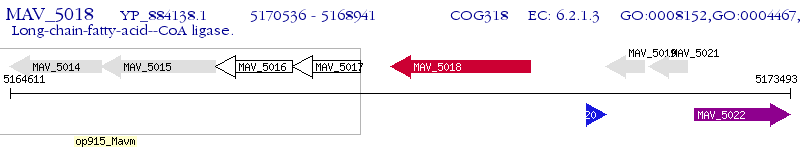
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5018 | - | - | 100% (531) | acyl-CoA synthetase |
| M. avium 104 | MAV_3523 | - | 2e-77 | 34.94% (518) | acyl-CoA synthase |
| M. avium 104 | MAV_1899 | - | 4e-69 | 35.78% (517) | feruloyl-CoA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0172 | fadD5 | 0.0 | 82.83% (530) | acyl-CoA synthetase |
| M. gilvum PYR-GCK | Mflv_0714 | - | 0.0 | 76.05% (522) | acyl-CoA synthetase |
| M. tuberculosis H37Rv | Rv0166 | fadD5 | 0.0 | 83.02% (530) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_02546 | fadD2 | 2e-41 | 27.78% (504) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_4569c | - | 0.0 | 74.61% (512) | fatty-acid-CoA ligase FadD |
| M. marinum M | MMAR_0409 | fadD5 | 0.0 | 82.67% (531) | fatty-acid-CoA ligase FadD5 |
| M. smegmatis MC2 155 | MSMEG_0131 | - | 0.0 | 75.10% (526) | acyl-CoA synthetase |
| M. thermoresistible (build 8) | TH_2878 | - | 0.0 | 75.00% (520) | PUTATIVE AMP-binding enzyme, putative |
| M. ulcerans Agy99 | MUL_0650 | fadD8 | 1e-56 | 32.50% (523) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_0131 | - | 0.0 | 78.63% (524) | acyl-CoA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0172|M.bovis_AF2122/97 -------------------MTAQLASHLTRALTLAQQQPYLARRQNWVNQ
Rv0166|M.tuberculosis_H37Rv -------------------MTAQLASHLTRALTLAQQQPYLARRQNWVNQ
MMAR_0409|M.marinum_M -------------------MTAELASHLPQAA-HALEQPYLARRQNWVNQ
MAV_5018|M.avium_104 --------------------MLSLTAQLAHHPTQANEQPYLSRRQNWVNQ
Mflv_0714|M.gilvum_PYR-GCK MTRTTSCLIKCHTSANPSTMERKTLTDPTGLSGSATEQPNLARRRNWANQ
Mvan_0131|M.vanbaalenii_PYR-1 -------MFDQISSFRNPSMEQKTLTEAIQQPGFATEQPYLARRQNWTNQ
MSMEG_0131|M.smegmatis_MC2_155 ----------------------------MTAEPSQIEQPYLARRQNWVNQ
TH_2878|M.thermoresistible__bu -----------------------------------LQQPYLARRQNWVNH
MAB_4569c|M.abscessus_ATCC_199 ----------------------MTDTIDATVQSAGLAQPYRARRQNWCNQ
MUL_0650|M.ulcerans_Agy99 ------------------------------------MSDDLLRNPTHNGH
MLBr_02546|M.leprae_Br4923 MTKLRHYLERGAAEVHYLRSIFESGALGFEPPLTYAAMAVAMRKWGDLAM
*.
Mb0172|M.bovis_AF2122/97 LERHAMMQPDAPALRFVG-NTMTWADLRRRVAALAGALSGRGVGFGDRVM
Rv0166|M.tuberculosis_H37Rv LERHAMMQPDAPALRFVG-NTMTWADLRRRVAALAGALSGRGVGFGDRVM
MMAR_0409|M.marinum_M LERHAMMQPRATAIRYLG-HTVTWADLRYRVAALAGSLSRRGVGFGDRVM
MAV_5018|M.avium_104 LERHALMQPNATALRFLG-KGLTWGELHGRVRALADALSRRGVGFGDRVM
Mflv_0714|M.gilvum_PYR-GCK LSRHALMQPDTVALRFLG-RSTTWGELDRRVTALAGALHERGVGFGDRVL
Mvan_0131|M.vanbaalenii_PYR-1 LARHALMQPDQPALRFLG-NTTTWGELDRRVSALAGALHKRGVGFGDRVL
MSMEG_0131|M.smegmatis_MC2_155 LTRHAMMQPDKTALRFLG-RTTTWRELDDRVTRLAGALNRRGVGFGDRVL
TH_2878|M.thermoresistible__bu LARHALMQPDATALRFMG-QTTTWADLHHRVNRLAGALRRRGVEFGDRVL
MAB_4569c|M.abscessus_ATCC_199 LARHALMQPDATAIRYLG-HSISWAELHQRVQALADALSRRGVGFGDRVL
MUL_0650|M.ulcerans_Agy99 LLVGALERHKNRPVLFLGDTTLTGGQLAERISQYIQAFEALGAGTGVTVG
MLBr_02546|M.leprae_Br4923 LPAMPARRSPHRAALIDEEGTLTYKELDRAAHALANGLIAKGVRGGDGIA
* . : . : :* .: *. * :
Mb0172|M.bovis_AF2122/97 ILMLNRTEFVESVLAANMIGAIAVPLNFRLTPTEIAVLVEDCAAHVMLTE
Rv0166|M.tuberculosis_H37Rv ILMLNRTEFVESVLAANMIGAIAVPLNFRLTPTEIAVLVEDCVAHVMLTE
MMAR_0409|M.marinum_M ILMLNRTEFVESVLAANMLGAIAVPLNFRLTPSEIAFLVEDCAPRLVITE
MAV_5018|M.avium_104 VLMLNRPEFMESVLAINMLGAIAVPLNFRLTAAEIAFLVQDCQARVVITE
Mflv_0714|M.gilvum_PYR-GCK ILMLNRTEFMESVLAVNKLGGIAVPVNFRMTPSEIAFLVSDCAAEVVITE
Mvan_0131|M.vanbaalenii_PYR-1 ILMLNRTEFIEAVLATNKLGAIAVPVNFRMTPAEIAFLVSDCEAKAIITE
MSMEG_0131|M.smegmatis_MC2_155 ILMLNRTEFVEAMLAANRLGAIAVPVNFRMTPPEIAYLVSDCEAHVVVTE
TH_2878|M.thermoresistible__bu ILMLNRTEFIETMLAANRLGAIAVPVNFRMTPPEIAFLAKDCEAKVIATE
MAB_4569c|M.abscessus_ATCC_199 ILMLNRPEYVESWLAINELGAIAIPVNFRMTPPEVAFLVENSGAKVAITE
MUL_0650|M.ulcerans_Agy99 LLSLNRPEVLMILGASQARGYRRTALHPLGSLDDHAYVLYDAGASALIID
MLBr_02546|M.leprae_Br4923 ILARNHRWFVIANYGCARVGARIILLNSEFSGPQIKEVSEREGAKVIIYD
:* *: : . * :: : : : . :
Mb0172|M.bovis_AF2122/97 A--ALAPVAIGVRNIQPLLSVIVVAG---GSSQDSVFGYEDLLNEAG-DV
Rv0166|M.tuberculosis_H37Rv A--ALAPVAIGVRNIQPLLSVIVVAG---GSSQDSVFGYEDLLNEAG-DV
MMAR_0409|M.marinum_M E--VLAQVAVGVREIAPALSTIVVAG---GASDPTVVGYDELISETG-DP
MAV_5018|M.avium_104 A--VLAPVATGVRDIESLLDTVVVAG---GSSDDAVLGYDDLIDETG-AA
Mflv_0714|M.gilvum_PYR-GCK S--VLENVATAVRDIDPKLATVIVAG---GTSRDNVLDYDDVLASTT-AA
Mvan_0131|M.vanbaalenii_PYR-1 A--VLANVATAVRDIDPTLATVISAG---GSG-DDVLDYDDVLAENA-PC
MSMEG_0131|M.smegmatis_MC2_155 P--VLAGVATAVRDLDATLQTVIVAGGATEDG---VLGYDDLIAEEG-EP
TH_2878|M.thermoresistible__bu P--VLAGVATAVRDLAPSLSSVIVAGAADGSDTPDVLDYEALIAEPG-ED
MAB_4569c|M.abscessus_ATCC_199 T--VLAPVAAAVRQQAPALETVIVAG---SASENGVLGYEEVIAEPG-EP
MUL_0650|M.ulcerans_Agy99 PNPMFVERALGLLEKVDSLEQVLTIGPVPEALEDRAVDLSAEAAKYQPKP
MLBr_02546|M.leprae_Br4923 D-----EYTKVVSKAEPALGKLRALGVNPDEEEPPGSTDETLAELIARSS
: : . * : * .
Mb0172|M.bovis_AF2122/97 HEPVDIPNDSPALIMYTAGTTGRPKGAVLTHANLTGQAMTALYTSGANIN
Rv0166|M.tuberculosis_H37Rv HEPVDIPNDSPALIMYTSGTTGRPKGAVLTHANLTGQAMTALYTSGANIN
MMAR_0409|M.marinum_M PEPVDIPNDSPALIMYTSGTTGRPKGAVLTHTNLTGQVMTALYTGGANIN
MAV_5018|M.avium_104 HQPVDIPNDAAALIMYTSGTTGRPKGAVLTHTNLTGQTMTGLYTNGADIN
Mflv_0714|M.gilvum_PYR-GCK PPLVDIPDDSPALIMYTSGTTGRPKGAVLTHNNLAGQAMTLLFTNGADIN
Mvan_0131|M.vanbaalenii_PYR-1 P-VVDIPNDAPALIMYTSGTTGRPKGAVLTHTNLTGQVMTLLFTNGADIN
MSMEG_0131|M.smegmatis_MC2_155 PAPVDIPNDSPALIMYTSGTTGRPKGAVLTHTNLSAQAMMHLFTTGADLN
TH_2878|M.thermoresistible__bu PPPVDIPDDSPALIMYTSGTTGNPKGAVLTHSNLTGQAMTNLFTIGVDLA
MAB_4569c|M.abscessus_ATCC_199 HAEVDLPGDTPALIMYTSGTTGRPKGAVLTHLNLSGQAMTYLLSTTVDLN
MUL_0650|M.ulcerans_Agy99 LVVADLPPNHIGGLTYTGGTTGKPKGVIGTAGNIT--AMTSIQLAEWEWP
MLBr_02546|M.leprae_Br4923 SAPAPKVRKRASIIILTSGTTGTPKGANR-NTPASLAPLGGILSKVPFRA
. . . : *.**** ***. : : :
Mb0172|M.bovis_AF2122/97 SDVGFVGVPLFHIAGIG-NMLTGLLLGLPTVIYPLGAFDPGQLLDVLEAE
Rv0166|M.tuberculosis_H37Rv SDVGFVGVPLFHIAGIG-NMLTGLLLGLPTVIYPLGAFDPGQLLDVLEAE
MMAR_0409|M.marinum_M SDVGFIGVPFFHIAGIG-NMLSGMLLGVPTVIYPLGAFNPGQLLDVLEAE
MAV_5018|M.avium_104 NDVGFIGVPFFHIAGIG-NMLTGLLLGIPTVIYPLGAFEPGQLLDVLAAE
Mflv_0714|M.gilvum_PYR-GCK HDIGFIGVPFFHIAGVG-SIVSGMMLGRPTVIYPLGAFDPNELLDVLEAE
Mvan_0131|M.vanbaalenii_PYR-1 HDIGFIGVPFFHIAGVG-SSVSGLLLGRPTVLYPVGAFDPNELLDVLEAE
MSMEG_0131|M.smegmatis_MC2_155 SDVGFVGVPLFHIAGVAGSVLSGVTLGVPTVLYPLGAFDPDGLLDVLESE
TH_2878|M.thermoresistible__bu NDVGFIGVPLFHIAGIAGNVLSGLTLGLPTVLYPLGAFDPGGLLDVLEAE
MAB_4569c|M.abscessus_ATCC_199 SDVGFIGVPMFHIAGVG-NMITGLLLGLPTVIHPLGAFDPGALLDVLESE
MUL_0650|M.ulcerans_Agy99 EHPRFLMCTPLSHAGAAFFTPTLVKGGE---MIVLAKFDPGEVLRVIEEQ
MLBr_02546|M.leprae_Br4923 HEVTLLPAPMFHALGYLHATLAMFLGST---LVLRRRFKPATVLEDIEKH
. :: . : * : . . : *.* :* : .
Mb0172|M.bovis_AF2122/97 KVTGIFLVPAQWQAVCTEQQ--ARPRDLR-LRVLSWGAAPAPDALLRQMS
Rv0166|M.tuberculosis_H37Rv KVTGIFLVPAQWQAVCTEQQ--ARPRDLR-LRVLSWGAAPAPDALLRQMS
MMAR_0409|M.marinum_M RVSGIFLVPAQWQAVCAEQQ--ARPRELS-LRVMSWGAAPAPDALLRQMS
MAV_5018|M.avium_104 KVTGIFLVPAQWQAVCAEQR--ARPRDLK-LRVISWGAAPAPDALLREMS
Mflv_0714|M.gilvum_PYR-GCK KVTGIFLVPAQWQAVCAAQK--ANPRNLK-LRFLSWGAAPASDTLLRQMS
Mvan_0131|M.vanbaalenii_PYR-1 KVTGIFLVPAQWQAVCAAQK--ARPRDLK-LRFLSWGAAPASDTLLREMA
MSMEG_0131|M.smegmatis_MC2_155 QVTGIFLVPAQWQAVCAAQR--ARPRNLK-LRALSWGAAPASDTLLREMN
TH_2878|M.thermoresistible__bu KVTGIFLVPAQWQAVCAEQR--ARPRDLR-LRALSWGAAPASDTLLQEMT
MAB_4569c|M.abscessus_ATCC_199 GVTGIFLVPAQWQAVCAAQQ--ANPRKVR-LKTLSWGAAPATDVLLRTMS
MUL_0650|M.ulcerans_Agy99 RITATMLVPSMLYALMDHPD--PHTRDLSSLETVYYGASAINPVRLAEAI
MLBr_02546|M.leprae_Br4923 QATAMVVVPVMLSRILDTLENLETKSDLSSLRVVFVSGSQLGAELATRAL
:. .:** : .: *. : ..:
Mb0172|M.bovis_AF2122/97 ATFPETQILAAFGQTEMSPVTCMLLGEDAIAKRGSVGRVIPTVAARVVDQ
Rv0166|M.tuberculosis_H37Rv ATFPETQILAAFGQTEMSPVTCMLLGEDAIAKRGSVGRVIPTVAARVVDQ
MMAR_0409|M.marinum_M EVFPGTQIMAAFGQTEMSPVTCMLLGEDAIRKRGSVGKVIPTVSARVVDD
MAV_5018|M.avium_104 AMFPGTQILAAFGQTEMSPVTCMLLGEDAIRKRGSVGKVIPTVAARVVDE
Mflv_0714|M.gilvum_PYR-GCK ETFPGAQILAAFGQTEMSPVTCMLLGDDALRKLGSVGKVIPTVSARIVDE
Mvan_0131|M.vanbaalenii_PYR-1 ETFPGAQILAAFGQTEMSPVTCMLLGDDAIRKLGSVGKVIPTVSARIVDE
MSMEG_0131|M.smegmatis_MC2_155 EVFPDAQIFAAFGQTEMSPVTCMLLGDDAIRKLGSVGRVIPTVSARVVDD
TH_2878|M.thermoresistible__bu EVFPGTKIFAAFGQTEMSPVTCMLLAEDAVRKLGSVGKVIPTVAARIVDD
MAB_4569c|M.abscessus_ATCC_199 ETFPDARILAAFGQTEMSPVTCMLMGDDAIRKMGSVGRVIPTVSARVVDD
MUL_0650|M.ulcerans_Agy99 DRFGPIFAQYYGQSEAPMVITYLPKADHDEKRLTSCGRPTLFARVALLGE
MLBr_02546|M.leprae_Br4923 EELG-PVIYNMYGSTEIAFATIAGPKDLQINS-ATVGPVVKGVKVKILDD
: . * : : * . . ::.:
Mb0172|M.bovis_AF2122/97 NMNDVPVGEVGEIVYRAPTLMSCYWNNPEATAEAFAGGWFHSGDLVRMDS
Rv0166|M.tuberculosis_H37Rv NMNDVPVGEVGEIVYRAPTLMSCYWNNPEATAEAFAGGWFHSGDLVRMDS
MMAR_0409|M.marinum_M EMNDVPIGQVGEIVYRAPTLMSGYWNNPDATAEAFAGGWFHSGDLVRMDE
MAV_5018|M.avium_104 NMNDVPVGEVGEIVYRAPTLMSGYWNNPEATAEAFAGGWFHSGDLVRMDE
Mflv_0714|M.gilvum_PYR-GCK DMNDVPIGQVGEIVYRAPTLMTGYWNNPTATAEAFAGGWFHSGDLVRQDE
Mvan_0131|M.vanbaalenii_PYR-1 DMNDVPVGQVGEIVYRAPTLMAGYWNNPKATAEAFAGGWFHSGDLVRQDE
MSMEG_0131|M.smegmatis_MC2_155 NMNDVPVGEVGEIVYRGPNLMVGYWNNPKATAEAFAGGWFHSGDLVRQDE
TH_2878|M.thermoresistible__bu TMNDVPVGEVGEIVYRAPTLMAGYWNNPQATAEAFAGGWFHSGDLVRQDE
MAB_4569c|M.abscessus_ATCC_199 NMNDVPVGEVGEIVYRAPTLMQGYWNNPEATADAFAGGWFHSGDLVRQDE
MUL_0650|M.ulcerans_Agy99 DGKPVPQGEPGEICVSGPLLAGGYWNKPDDTAQTFRDGWLHTGDMAREDE
MLBr_02546|M.leprae_Br4923 SGKEVPRGQVGRIFVGNAFPFEGYTG---GGGKQIIDGLLSSGDVGYFDE
: ** *: *.* . * . .. : .* : :**: *.
Mb0172|M.bovis_AF2122/97 DGYVWVVDRKKDMIISGGENIYCAELENVLASHPDIAEVAVIGRADEKWG
Rv0166|M.tuberculosis_H37Rv DGYVWVVDRKKDMIISGGENIYCAELENVLASHPDIAEVAVIGRADEKWG
MMAR_0409|M.marinum_M DGYVWVVDRKKDMIISGGENVYCAEVENVLASHPSIVEVAVIGRADEKWG
MAV_5018|M.avium_104 DGYVWVVDRKKDMIISGGENIYCAEVENVLASHPDIVEVAVIGRAHEKWG
Mflv_0714|M.gilvum_PYR-GCK EGYIWVVDRKKDMIISGGENIYCAEVENVLASHPDIVEVAVIGRSHRKWG
Mvan_0131|M.vanbaalenii_PYR-1 EGYIWVVDRKKDMIISGGENIYCAEVENVLAAHPEIVEVAVIGRAHEKWG
MSMEG_0131|M.smegmatis_MC2_155 EGYVWVVDRKKDMIISGGENIYCAEVENVLAAHPAIMEVAVIGRPHEKWG
TH_2878|M.thermoresistible__bu EGYIWVVDRKKDMIISGGENIYCAEVENVLAAHPAVAEVAVVGRPHEKWG
MAB_4569c|M.abscessus_ATCC_199 DGFVWVVDRKKDMIISGGENIYCAEVENALAEHEQIAEVAVIGRPHEKWG
MUL_0650|M.ulcerans_Agy99 DGFYFIVDRVKDMIVTGGFNVFPREVEDVVAEHSAVAQVCVVGAPDEKWG
MLBr_02546|M.leprae_Br4923 HDLLYISGRDDEMIVSGGENVFPAEVEDLISGHPEVVEATAIGVDDKEWG
.. :: .* .:**::** *:: *:*: :: * : :. .:* ..:**
Mb0172|M.bovis_AF2122/97 EVPIAVAAVTN--------DDLRIEDLGEFLTDRLARYKHPKALEIVDAL
Rv0166|M.tuberculosis_H37Rv EVPIAVAAVTN--------DDLRIEDLGEFLTDRLARYKHPKALEIVDAL
MMAR_0409|M.marinum_M EVPIAVAAVTK--------DHLRIEDLDEYLTERLARYKHPKALEIVDAL
MAV_5018|M.avium_104 EVPIAVAAVAN--------DNLALEDLDEFLTERLARYKHPKALEIVDAL
Mflv_0714|M.gilvum_PYR-GCK EVPVAVAVLRSSLN-STGSGRLELTDLEQFLGDRLARYKHPKAIEIVEAL
Mvan_0131|M.vanbaalenii_PYR-1 EVPVAVAVVRSSLE-ATPGAGIELADLEQFLTERLARYKHPKALEIVDAL
MSMEG_0131|M.smegmatis_MC2_155 EVPVAVVALNS-----ATAESISLGELDSFLTERLARYKHPKALEVVDAL
TH_2878|M.thermoresistible__bu EVPVAVVALN-------PDAELTMAELEEFLADRLARYKHPKVLEIVDAL
MAB_4569c|M.abscessus_ATCC_199 EVPVAVAAIHG--------AALTLAELDGFLTARLARYKHPKDLVIVDAL
MUL_0650|M.ulcerans_Agy99 EAVTAVVVLRADAARDDAAIETMTAEIQAAVKQRKGSVQSPKQVVVVDSL
MLBr_02546|M.leprae_Br4923 ARLRAFVVKKQ-------DATIDEDAIKAYVRDHLARYKVPREVIFLEEL
*... : : : . : *: : .:: *
Mb0172|M.bovis_AF2122/97 PRNPAGKVLKTELRLRYGACVNVERRSASAGFTERRENRQKL
Rv0166|M.tuberculosis_H37Rv PRNPAGKVLKTELRLRYGACVNVERRSASAGFTERRENRQKL
MMAR_0409|M.marinum_M PRNPSGKVLKTELRLRYGVGVYSESRSVSRSFEFREGS----
MAV_5018|M.avium_104 PRNPAGKVLKTELRIRYGGG----------------------
Mflv_0714|M.gilvum_PYR-GCK PRNPAGKVLKTELRVRFGSEESVDAGESATAATSSAAAPKD-
Mvan_0131|M.vanbaalenii_PYR-1 PRNPAGKVLKTELRIRYSAASFADAGESSTPPTISTAPQRN-
MSMEG_0131|M.smegmatis_MC2_155 PRNPSGKVLKTELRERFGRATSLDGGESNSATTVSAG-----
TH_2878|M.thermoresistible__bu PRNPAGKVLKTELRARFAAGPSAEKEQSSSGATTSAGSS---
MAB_4569c|M.abscessus_ATCC_199 PRNPSGKVLKTELRQRFGIAD---------------------
MUL_0650|M.ulcerans_Agy99 PLTGLGKPDKKAVRARFWEGAGRAVG----------------
MLBr_02546|M.leprae_Br4923 PRNPTGKVLKRELRDFEVH-----------------------
* . ** * :*