For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LVTVFLVDDHEVVRRGLIDLLDADPQLDVVGEAGSVSEAMARIPAANPDVAVLDVRLPDGDGIELCRDLL SRMPNLRCLILTSYTSDEAMLDAILAGASGYVVKDIKGMELARAIKEVGAGRSLLDNRAAAALMSRLRGV VERPDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNYVSRLLAKLGMERRTQAAVFAAELKRA RSAGHR
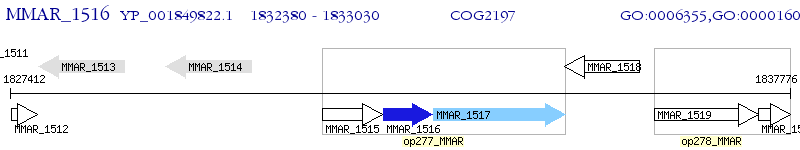
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1516 | devR | - | 100% (216) | two-component transcriptional regulatory protein DevR |
| M. marinum M | MMAR_3480 | - | 5e-95 | 84.54% (207) | two-component transcriptional regulatory protein |
| M. marinum M | MMAR_3959 | - | 4e-29 | 34.30% (207) | two-component transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3157c | devR | 1e-105 | 89.72% (214) | two component transcriptional regulatory protein DevR |
| M. gilvum PYR-GCK | Mflv_3847 | - | 2e-97 | 84.19% (215) | two component LuxR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3133c | devR | 1e-105 | 89.72% (214) | two component transcriptional regulatory protein DevR |
| M. leprae Br4923 | MLBr_01286 | - | 1e-10 | 30.82% (159) | putative two-component response regulatory protein |
| M. abscessus ATCC 19977 | MAB_3891c | - | 4e-84 | 69.77% (215) | LuxR family transcriptional regulator |
| M. avium 104 | MAV_4109 | - | 2e-93 | 82.30% (209) | two component transcriptional regulatory protein devr |
| M. smegmatis MC2 155 | MSMEG_5244 | - | 1e-96 | 85.65% (209) | two component transcriptional regulatory protein devr |
| M. thermoresistible (build 8) | TH_2159 | - | 4e-96 | 83.25% (209) | TWO COMPONENT TRANSCRIPTIONAL REGULATORY PROTEIN DEVR (PROBABLY |
| M. ulcerans Agy99 | MUL_2423 | devR | 1e-116 | 100.00% (216) | two component transcriptional regulatory protein DevR |
| M. vanbaalenii PYR-1 | Mvan_2548 | - | 2e-97 | 86.12% (209) | two component LuxR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1516|M.marinum_M ------------MVTVFLVDDHEVVRRGLIDLLDADPQLDVVGEAGSVSE
MUL_2423|M.ulcerans_Agy99 ------------MVTVFLVDDHEVVRRGLIDLLDADPQLDVVGEAGSVSE
Mb3157c|M.bovis_AF2122/97 ------------MVKVFLVDDHEVVRRGLVDLLGADPELDVVGEAGSVAE
Rv3133c|M.tuberculosis_H37Rv ------------MVKVFLVDDHEVVRRGLVDLLGADPELDVVGEAGSVAE
Mflv_3847|M.gilvum_PYR-GCK ------------MVKVFLVDDHEVVRRGLIDLLGSDPELDVVGEAGSVSE
Mvan_2548|M.vanbaalenii_PYR-1 ------------MVKVFLVDDHEVVRRGLIDLLSSDPELDVVGEAGSVAE
MSMEG_5244|M.smegmatis_MC2_155 ------------MIRVFLVDDHEVVRRGLIDLLSADPELDVIGEADSVSQ
TH_2159|M.thermoresistible__bu ------------MVSVFLVDDHEVVRRGLIDLLDSDPDLEVVGEAGTVAQ
MAV_4109|M.avium_104 ------------MVKVFLVDDHEVVRRGLCDLLSSDPDLRIVGEAGTVAE
MAB_3891c|M.abscessus_ATCC_199 ------------MITVFLVDDHEVVRRGLADLLEEEADFSVIGQASSVAE
MLBr_01286|M.leprae_Br4923 MTGPTTDADAATPHRVLIAEDEALIRLDLAEMLR-DEGYDIVGEAADGQE
*::.:*. ::* .* ::* : ::*:* :
MMAR_1516|M.marinum_M AMARIPAANPDVAVLDVRLPDGDGIELCRDLLSRMPNLRCLILTSYTSDE
MUL_2423|M.ulcerans_Agy99 AMARIPAANPDVAVLDVRLPDGDGIELCRDLLSRMPNLRCLILTSYTSDE
Mb3157c|M.bovis_AF2122/97 AMARVPAARPDVAVLDVRLPDGNGIELCRDLLSRMPDLRCLILTSYTSDE
Rv3133c|M.tuberculosis_H37Rv AMARVPAARPDVAVLDVRLPDGNGIELCRDLLSRMPDLRCLILTSYTSDE
Mflv_3847|M.gilvum_PYR-GCK ALTRVPTAQPDVAVLDVRLPDGNGIELCRELLSLMPDLRCLMLTSFTSDE
Mvan_2548|M.vanbaalenii_PYR-1 ALARIPALQPDVAVLDVRLPDGNGIELCRDLLSRLPDLRCLMLTSFTSDE
MSMEG_5244|M.smegmatis_MC2_155 ALARIPAAQPDVAVLDVRLPDGNGIELCRDLLSHMPNLRCLMLTSFTSDE
TH_2159|M.thermoresistible__bu AMARIPAAQPDVAVLDVRLPDGNGVELCRDLLARMPDLRCLMLTSYTSDQ
MAV_4109|M.avium_104 AKARIPAARPDVAVLDVRLPDGNGIELCRDLLSEHPDLRCLMLTSFTSDE
MAB_3891c|M.abscessus_ATCC_199 AMARIPALQPDIAVLDIRLPDGNGVELCRELRSKLPNLNCLMLTSFTDEH
MLBr_01286|M.leprae_Br4923 AVELAERHKPDLVIMDVKMPRRDGIDAASEIASKRIAP-IVVLTAFSQRD
* .**:.::*:::* :*:: . :: : ::**:::. .
MMAR_1516|M.marinum_M AMLDAILAGASGYVVKDIKGMELARAIKEVGAGRSLLDNRAAAALMSRLR
MUL_2423|M.ulcerans_Agy99 AMLDAILAGASGYVVKDIKGMELARAIKEVGAGRSLLDNRAAAALMSRLR
Mb3157c|M.bovis_AF2122/97 AMLDAILAGASGYVVKDIKGMELARAVKDVGAGRSLLDNRAAAALMAKLR
Rv3133c|M.tuberculosis_H37Rv AMLDAILAGASGYVVKDIKGMELARAVKDVGAGRSLLDNRAAAALMAKLR
Mflv_3847|M.gilvum_PYR-GCK AMLDAILAGASGYVVKDIKGMELARAIKEVGSGRSLLDNRAAAALMAKLR
Mvan_2548|M.vanbaalenii_PYR-1 AMLDAILAGASGYVVKDIKGMELAQAIKDVGSGRSLLDNRAAAALMARLR
MSMEG_5244|M.smegmatis_MC2_155 AMLDAILAGASGYVVKDIKGMELAQAIKDVGAGKSLLDNRAATALMSKLR
TH_2159|M.thermoresistible__bu AMLDAILAGASGYVVKDIKGMELAQAIKDVGAGRSLLDNRAAEALMAKLR
MAV_4109|M.avium_104 AMLEAILAGASGFVIKDIKGMELARAIKDVGAGKSLLDNRAAAALMAKLR
MAB_3891c|M.abscessus_ATCC_199 AMLDAIMAGAGGYVIKDIKGMELISAVRTVGAGRSLLDNRAAAALMNRLR
MLBr_01286|M.leprae_Br4923 LVERARDAGAMAYLVKPFTISDLIPAIALAMSRFSELT-----ALEREVA
: * *** .:::* :. :* *: . : * * ** .:
MMAR_1516|M.marinum_M GVVERPDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNYVSRL
MUL_2423|M.ulcerans_Agy99 GVVERPDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNYVSRL
Mb3157c|M.bovis_AF2122/97 GAAEKQDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNYVSRL
Rv3133c|M.tuberculosis_H37Rv GAAEKQDPLSGLTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNYVSRL
Mflv_3847|M.gilvum_PYR-GCK GEGERTDPLSGLTDQERVLLDLLGEGLTNKQIAARMFLAEKTVKNYVSRL
Mvan_2548|M.vanbaalenii_PYR-1 GEGERTDPLSGLTEQERVLLGLLGEGLTNKQIAARMFLAEKTVKNYVSRL
MSMEG_5244|M.smegmatis_MC2_155 GDAERSDPLSGLTQQERVLLDLLGEGLTNKQIAARMFLAEKTVKNYVSRL
TH_2159|M.thermoresistible__bu GADHQPDPLAGLSDQERVLLSLIGEGLTNKQIAARMFLAEKTVKNYVSRL
MAV_4109|M.avium_104 GSAAQADPLSGLTEQERTLLGLLSEGLTNRQIAARMFLAEKTVKNYVSRL
MAB_3891c|M.abscessus_ATCC_199 AAADNPSPLAGLTAQERTLLELIGEGLTNRQIAERMFLAEKTVKNYVSRL
MLBr_01286|M.leprae_Br4923 TLADRLETRK-LVERAKGLLQVKQG------------MTEPEAFKWIQRA
. .. * : : ** : ::* . :::.*
MMAR_1516|M.marinum_M LAKLGMERRTQAAVFAAELKRARSAGHR------
MUL_2423|M.ulcerans_Agy99 LAKLGMERRTQAAVFAAELKRARSAGHR------
Mb3157c|M.bovis_AF2122/97 LAKLGMERRTQAAVFATELKRSRPPGDGP-----
Rv3133c|M.tuberculosis_H37Rv LAKLGMERRTQAAVFATELKRSRPPGDGP-----
Mflv_3847|M.gilvum_PYR-GCK LAKLGMERRTQAAVFISKLDRGHGAGNA------
Mvan_2548|M.vanbaalenii_PYR-1 LAKLGMERRTQAAVFVSKLERHHRPDGE------
MSMEG_5244|M.smegmatis_MC2_155 LAKLGMERRTQAAVFASKLDRRN-----------
TH_2159|M.thermoresistible__bu LAKLGMERRTQAAVFVSKLDRPIDPRG-------
MAV_4109|M.avium_104 LAKLGMERRTQAAVFASKLNQQAGRPPTPPDWPG
MAB_3891c|M.abscessus_ATCC_199 LTKLDLERRTQVAVLATKLARENQGLHG------
MLBr_01286|M.leprae_Br4923 AMDRRTTMKRVAEVVLETLDTPKDT---------
. : . *. *