For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLLNPNRLQRKYPDARSAEIMAATVDFFESRGKARLKRDDHDRAWYSDFLNHIAEHRIFASLLTPSEYGA DDCRWDTYRISEFAEIIGFYGLSYWYPFQVTALGLGPIWMSDNEDAKRKAAAQLEQGEVFAFGLSEQAHG ADVYQTDMILTPSEHGWTASGEKYYIGNANVARMVSTFGKIEDSSKEPEYVFFAADSQHDRYELKKNVVN SQNYVANYALHDYPVTEADLLHRGMGAFHAALNTVNVCKYNLGWGAVGMCTHAFYEAITHAANRHLYGTV VTDFSHVRRLLTDAYARLVAMRLVASRASDYMRSASAEDRRYLLYSPLTKAKVTSEGERVIAALWDVIAA KGVEKDTFFETVAREIGLLPRLEGTVHINIGLLAKFMPNFLFVPDAALPLIPRRDDAADDTFLFNQGPTG GLGKVRFHDWRAPFDSYGHLPNVALLREQIDVLTEMLAGATPDASQQKDIDFAFGVGQIFALVPYAQLIL EEAPLSGVDDALLDEIFGLLVRDFNSYAVELNDKAATTDQQAQFAMRMIRRPAHDAARYDQVWKEHVLPL NGAYEMRP
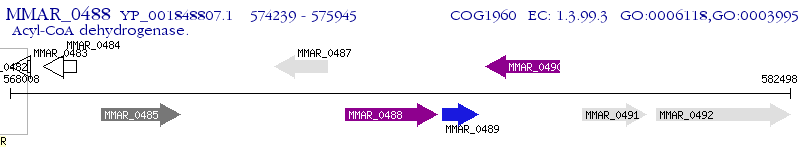
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0488 | fadE4 | - | 100% (568) | acyl-CoA dehydrogenase FadE4 |
| M. marinum M | MMAR_0698 | fadE7 | 2e-07 | 22.22% (198) | acyl-CoA dehydrogenase FadE7 |
| M. marinum M | MMAR_3847 | fadE19 | 9e-06 | 21.94% (319) | acyl-CoA dehydrogenase FadE19 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0236 | fadE4 | 0.0 | 84.51% (568) | acyl-CoA dehydrogenase FADE4 |
| M. gilvum PYR-GCK | Mflv_0398 | - | 0.0 | 55.73% (576) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv0231 | fadE4 | 0.0 | 84.51% (568) | acyl-CoA dehydrogenase FADE4 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3481 | - | 1e-160 | 50.26% (573) | acyl-CoA dehydrogenase FadE |
| M. avium 104 | MAV_4939 | - | 0.0 | 82.60% (569) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1821 | - | 3e-07 | 22.19% (329) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4603 | - | 6e-13 | 21.15% (383) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_1151 | fadE4 | 0.0 | 99.65% (568) | acyl-CoA dehydrogenase FadE4 |
| M. vanbaalenii PYR-1 | Mvan_0281 | - | 1e-178 | 54.17% (587) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0398|M.gilvum_PYR-GCK ------MAARGLLLNPNHFDPAWLDPESRRLLRALIDWFEQRGKARLLRD
Mvan_0281|M.vanbaalenii_PYR-1 ------MPETPLLLDPNNFDPKRLDPESRRLLAALIDWFEQRGKVRLLRD
MAB_3481|M.abscessus_ATCC_1997 MSQDTNAAPKTELLHPTADDFAEFDGETRRLLKATVDWFEERGKTRLLQD
MMAR_0488|M.marinum_M -----------MLLNPNRLQRKYPDARSAEIMAATVDFFESRGKARLKRD
MUL_1151|M.ulcerans_Agy99 -----------MLLNPNRLQRKYPDARSAEIMAATVDFFESRGKARLKRD
Mb0236|M.bovis_AF2122/97 -----------MLLNPNHLTRKYPDRRSGEIMAATVDFFESRGKARLKHD
Rv0231|M.tuberculosis_H37Rv -----------MLLNPNHLTRKYPDRRSGEIMAATVDFFESRGKARLKHD
MAV_4939|M.avium_104 -----------MLLNPNHLQRRYPDRRSAEIMAATVDFFESRGKARLKSD
MSMEG_1821|M.smegmatis_MC2_155 ---------MAAWGGNPSFDLFQLPEEHQELRAAIRALAEKEIAPHAADV
TH_4603|M.thermoresistible__bu ----------------MAVDRLLPSQEAAELIELTREIADKVLDPIVDRH
. .: :.
Mflv_0398|M.gilvum_PYR-GCK DLDGVWVADFLEFVKKERLFATFLTPSEFAEGDPNKRWDTARNAVLSEIL
Mvan_0281|M.vanbaalenii_PYR-1 DLNAEWVTDFLDFVRRERLFATFLTPSEFANGDPNKRWDTSRNAVLSEIL
MAB_3481|M.abscessus_ATCC_1997 YVDKVFYQDFLDFAGREKLFATFLTPAADGAGNTEKRWDTARVAKLSEIL
MMAR_0488|M.marinum_M DHDRAWYSDFLNHIAEHRIFASLLTPSEYGADD--CRWDTYRISEFAEII
MUL_1151|M.ulcerans_Agy99 DHDRAWYSDFLNHIAEHRIFASLLTPSEYGADD--CRWDTYRISEFAEII
Mb0236|M.bovis_AF2122/97 DHERIWYSDFLDFVGRERIFASLLTPASYGADD--CRWDTYRISEFAEIM
Rv0231|M.tuberculosis_H37Rv DHERIWYSDFLDFVGRERIFASLLTPASYGADD--CRWDTYRISEFAEIM
MAV_4939|M.avium_104 DHERVWYSDFLEHIGRERIFASLLTPTRYGSAD--CRWDTYRISGFAEIL
MSMEG_1821|M.smegmatis_MC2_155 DENARFPQEALDALN-ASGFNAVHVPEEYGGQG----ADSVAACIVIEEV
TH_4603|M.thermoresistible__bu EKDETYPEGVFEQLG-AAGLLSLPQPEEWGGGG----QPYEVYLQVLEEI
: : :: : :. * . . . * :
Mflv_0398|M.gilvum_PYR-GCK GFYGLSYWYAEQVTILGLGPIWQSDNVKARERAAAQLEAGE-VMAFGLSE
Mvan_0281|M.vanbaalenii_PYR-1 GFYGLSYWYAEQVTILGLGPIWQSGNVKAKERAAEQLEAGG-VMAFGLSE
MAB_3481|M.abscessus_ATCC_1997 GFYGLNYWYPWQVTVLGLGPVWQSANTDPRKRAADLLDGGA-VAAFGLSE
MMAR_0488|M.marinum_M GFYGLSYWYPFQVTALGLGPIWMSDNEDAKRKAAAQLEQGE-VFAFGLSE
MUL_1151|M.ulcerans_Agy99 GFYGLSYWYPFQVTALGLGPIWMSDNEDAKRKAAAQLEQGE-VFAFGLSE
Mb0236|M.bovis_AF2122/97 GFYGLSYWYPFQVTALGLGPIWMSANEDAKRKAAAGLEAGE-VFAFGLSE
Rv0231|M.tuberculosis_H37Rv GFYGLSYWYPFQVTALGLGPIWMSANEDAKRKAAAGLEAGE-VFAFGLSE
MAV_4939|M.avium_104 GFYGLNYWYPFQVTALGLGPIWMSGNDEAKRKAAGLLEAGE-VFAFGLSE
MSMEG_1821|M.smegmatis_MC2_155 ARVDASASLIPAVNKLGTMGLILRGSDELKKQVLPQLASGAAMASYALSE
TH_4603|M.thermoresistible__bu AARWASVAVAVSVHSLSSHPLLVFGTEEQKKRWLPGMLSGEQIGAYSLSE
. . * *. : . . :.: : * : ::.***
Mflv_0398|M.gilvum_PYR-GCK RAHGADIYNTDMLLTPASP-----------GADDPVVFRASGEKYYIGNG
Mvan_0281|M.vanbaalenii_PYR-1 RDHGADIYNTDMLLTPAGDERSREEQTTADGDDDGVLFRASGEKYYIGNG
MAB_3481|M.abscessus_ATCC_1997 QAHGADIYSSDLVLTPVDS-------------SEGGGYKASGRKYYIGNG
MMAR_0488|M.marinum_M QAHGADVYQTDMILTPS-----------------EHGWTASGEKYYIGNA
MUL_1151|M.ulcerans_Agy99 QAHGADVYQTDMILTPS-----------------EHGWTASGEKYYIGNA
Mb0236|M.bovis_AF2122/97 QTHGADVYQTDMILTPS-----------------DGGWTANGEKYYIGNA
Rv0231|M.tuberculosis_H37Rv QTHGADVYQTDMILTPS-----------------DGGWTANGEKYYIGNA
MAV_4939|M.avium_104 QTHGADVYQTDMNLTPAP----------------GGGWTAAGQKYYIGNA
MSMEG_1821|M.smegmatis_MC2_155 REAGSDAASMRTRARAD-----------------GDDWVLNGTKCWITNG
TH_4603|M.thermoresistible__bu PQAGSDAAALRCAATPT-----------------DGGYVINGSKSWITHG
*:* . : * * :* :.
Mflv_0398|M.gilvum_PYR-GCK NVAGMVSVFGRRTDVEGADGYVWFVADSRHPDYELIGNVVHG---QMFVS
Mvan_0281|M.vanbaalenii_PYR-1 NVAGMVSVFARRTDVEGADGYVWFVADSGHERYELIGNVVHG---QMFVS
MAB_3481|M.abscessus_ATCC_1997 NCARIVSVFGRVQGRDGLDQYVFFLADSEHPNYRVIKNVVPS---QMYVA
MMAR_0488|M.marinum_M NVARMVSTFGKIEDSSKEPEYVFFAADSQHDRYELKKNVVNS---QNYVA
MUL_1151|M.ulcerans_Agy99 NVARMVSTFGKIEDSSKEPEYVFFAADSQHDRYELKKNVVNS---QNYVA
Mb0236|M.bovis_AF2122/97 NVARMVSTFGKIAGTPESQEYVFFVADSQHERYDLIKNVVNS---QNYVA
Rv0231|M.tuberculosis_H37Rv NVARMVSTFGKIAGTPESQEYVFFVADSQHERYDLIKNVVNS---QNYVA
MAV_4939|M.avium_104 NVARMVSTFGKFAGGATDSEYVFFAADSRHERYRLIKNVVNS---QNYVG
MSMEG_1821|M.smegmatis_MC2_155 GKSSWYTVMAVTDPDKGANGISAFMVHQDDEGFSVGPKERKLGIKGSPTT
TH_4603|M.thermoresistible__bu GKADFYTLFARTG--EGSRGVSCFLVPADQPGLSFGKPEEKMGLHAVPTT
. : : :. * . . . .
Mflv_0398|M.gilvum_PYR-GCK NFALRDYPVYEEDILATGQDAFSAALNTVNVGKFNLCSGSIGMCEHAFYE
Mvan_0281|M.vanbaalenii_PYR-1 NFALHDYPVHEEDILATGPDAFSAALNTVNVGKFNLCSGSIGMCEHAFYE
MAB_3481|M.abscessus_ATCC_1997 EFELDEYPVSEDDILHVGADAFSAALNTVNIGKFNLCFGGIGLSTHAFYE
MMAR_0488|M.marinum_M NYALHDYPVTEADLLHRGMGAFHAALNTVNVCKYNLGWGAVGMCTHAFYE
MUL_1151|M.ulcerans_Agy99 NYALHDYPVTEADLLHRGMGAFHAALNTVNVCKYNLGWGAVGMCTHAFYE
Mb0236|M.bovis_AF2122/97 NYALRDYPVTEADILHRGAEAFHAALNTVNVCKYNLGWGAIGMCTHALYE
Rv0231|M.tuberculosis_H37Rv NYALRDYPVTEADILHRGAEAFHAALNTVNVCKYNLGWGAIGMCTHALYE
MAV_4939|M.avium_104 NFALSDYPVTEADILHRGPEAFHAALNTVNVCKYNLGWGAVGMCTHAMYE
MSMEG_1821|M.smegmatis_MC2_155 ELYFENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDA
TH_4603|M.thermoresistible__bu SAFYDNARIDADRRIGEEGQGLQIAFSALDSGRLGIAAVATGLAQAALDE
. : : : .: *: ::: : : . *:. *:
Mflv_0398|M.gilvum_PYR-GCK AITHANNRILYGNPVTDFPHVRATFVDAYARLIAMKLFSDRAIDYFRSAG
Mvan_0281|M.vanbaalenii_PYR-1 AINHANNRILYGNPVTAFPHVRAGFVDAYTRLIAMKLFSDRAIDYFRSAG
MAB_3481|M.abscessus_ATCC_1997 SITHAHNRILYGNPVTKFDHVRREFVDAYARLLAMKLFSERAVDYFRTAS
MMAR_0488|M.marinum_M AITHAANRHLYGTVVTDFSHVRRLLTDAYARLVAMRLVASRASDYMRSAS
MUL_1151|M.ulcerans_Agy99 AITHAANRHLYGTVVTDFSHVRRLLTDAYARLVAMRLVASRASDYMRSAS
Mb0236|M.bovis_AF2122/97 SVTHAANRHLYGTVVTDFSHVRRLLTDAYVRLIAMKLVASRASDYMRSAS
Rv0231|M.tuberculosis_H37Rv SVTHAANRHLYGTVVTDFSHVRRLLTDAYVRLIAMKLVASRASDYMRSAS
MAV_4939|M.avium_104 AVTHASNRILYGTVVTDFSHVRRLLIDAYARLVAMRLVATRACDYMRSAS
MSMEG_1821|M.smegmatis_MC2_155 AIAYTKDRKQFGTSISDFQAVQFMLADMAMKVESARLMVYHAAA---RAE
TH_4603|M.thermoresistible__bu AVAYANERTAFGRKIIDHQGLGFLLADMAAAVATARATYLDAAR---RRD
:: :: :* :* : . : : * : : : *
Mflv_0398|M.gilvum_PYR-GCK PHDRRYLLFNPVTKSKVTSEGEKVVTLLWDVLAAKGFEKDTYFYQVTRLI
Mvan_0281|M.vanbaalenii_PYR-1 PDDRRYLLFNPVTKSKVTSEGEKVVTLLWDVLAAKGFEKNTYFYQVTRLI
MAB_3481|M.abscessus_ATCC_1997 ADDRRYLLFNPVTKMKATTEAEKVLALLADVMSAKAFEAESYTTAARIDV
MMAR_0488|M.marinum_M AEDRRYLLYSPLTKAKVTSEGERVIAALWDVIAAKGVEKDTFFETVAREI
MUL_1151|M.ulcerans_Agy99 AEDRRYLLYSPLTKAKVTSEGERVIAALWDVIAAKGVEKDTFFETVAREI
Mb0236|M.bovis_AF2122/97 AADRRYLLYSPLTKAKVTSEGERVITALWDVIAAKGVEKDTFFETVAREI
Rv0231|M.tuberculosis_H37Rv AADRRYLLYSPLTKAKVTSEGERVITALWDVIAAKGVEKDTFFETVAREI
MAV_4939|M.avium_104 ATDRRYLLYSPLTKAKITGEGERVITALWDVIAAKGVEKDTIFEAMTREI
MSMEG_1821|M.smegmatis_MC2_155 RGEKNLGFISAASKCLASDIAMEVTTDAVQLFGGAGYTQDFPVERMMRDA
TH_4603|M.thermoresistible__bu QG-RPYSQQASIAKLTATDAAMKVTTDAVQVFGGVGYTRDYRVERYMREA
: . :* : . .* : :::.. . :
Mflv_0398|M.gilvum_PYR-GCK GALPRLEGTVHVNVAQILKFMPNYMFNP-AEYPPIGTRDDPADDVFFWSQ
Mvan_0281|M.vanbaalenii_PYR-1 GALPRLEGTVHVNVAQILKFMPNFMFNP-APYPEIGTRDDPADDVFFWSQ
MAB_3481|M.abscessus_ATCC_1997 AGLPKLEGTVAVNLALILKFMPAYLHDA-QDLPVPGVQLQPGDDEFLFDQ
MMAR_0488|M.marinum_M GLLPRLEGTVHINIGLLAKFMPNFLFVPDAALPLIPRRDDAADDTFLFNQ
MUL_1151|M.ulcerans_Agy99 GLLPRLEGTVHINIGLLAKFMPNFLFAPDAALPLIPRRDDAADDTFLFNQ
Mb0236|M.bovis_AF2122/97 GLLPRLEGTVHINIGLLGKFMPNYLFAPDSTLPVIPRRDDAADDAFLFAQ
Rv0231|M.tuberculosis_H37Rv GLLPRLEGTVHINIGLLGKFMPNYLFAPDSTLPVIPRRDDAADDAFLFAQ
MAV_4939|M.avium_104 GLLPRLEGTVHINIGLLGKFMPNYLFAPSGELPVIGRRDDAADDEFLFAQ
MSMEG_1821|M.smegmatis_MC2_155 KITQIYEGTNQIQRVVMSRALLR---------------------------
TH_4603|M.thermoresistible__bu KIMQIFEGTNQIQRLVIARGLTR---------------------------
*** :: : : :
Mflv_0398|M.gilvum_PYR-GCK GPARGASKVRFADWAPVYERHAGIPNVGRFYEQVQAFKQLLTVSAPDADQ
Mvan_0281|M.vanbaalenii_PYR-1 GPARGASKVRFADWAPVYERNTGIPNVAVFYQQVQAFKDLLATAAPDADQ
MAB_3481|M.abscessus_ATCC_1997 GPASGLGKVRFHDWRKPFADNADIPNVAAFTQRAEALATLVANER-DAIK
MMAR_0488|M.marinum_M GPTGGLGKVRFHDWRAPFDSYGHLPNVALLREQIDVLTEMLAGATPDASQ
MUL_1151|M.ulcerans_Agy99 GPTGGLGKVRFHDWRAPFGSYGHLPNVALLREQIDVLTEMLAGATPDASQ
Mb0236|M.bovis_AF2122/97 GPTGGLGKVRFHDWRASFDTCAHLPNVALLREQVDVFAELLASATPDAAQ
Rv0231|M.tuberculosis_H37Rv GPTGGLGKVRFHDWRASFDTCAHLPNVALLREQVDVFAELLASATPDAAQ
MAV_4939|M.avium_104 GPTGGLGKVRFGDWRAPFDGFAHLPNVALLRAQIDVLAEMLASATPDAVQ
MSMEG_1821|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
Mflv_0398|M.gilvum_PYR-GCK QADLDFVLVVGHLFSLVVYGQLILEQAEITGLDRDVLDQIFDVAVRDFSA
Mvan_0281|M.vanbaalenii_PYR-1 QRDLDFVLVVGHLFTLVVYGQLILEQAELTGLESDLVDQIFDVQIRDFSA
MAB_3481|M.abscessus_ATCC_1997 NAGLDLQLSIGELFTLVVYGGLILEQAQLRGTDSALIDQIFEFLNRDFSV
MMAR_0488|M.marinum_M QKDIDFAFGVGQIFALVPYAQLILEEAPLSGVDDALLDEIFGLLVRDFNS
MUL_1151|M.ulcerans_Agy99 QKDIDFAFGVGQIFALVPYAQLILEEAPLSGVDDALLDEIFGLLVRDFNS
Mb0236|M.bovis_AF2122/97 QKDIDFAFGVGQLFANVPYAQLILEEARLSGVDEALIDEIFGVLVRDFNT
Rv0231|M.tuberculosis_H37Rv QKDIDFAFGVGQLFANVPYAQLILEEARLSGVDEALIDEIFGVLVRDFNT
MAV_4939|M.avium_104 QKDIDFAFAVGQLFATVPYAQLILEEATLSGVDDALIDEIFGVLVRDFNA
MSMEG_1821|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
Mflv_0398|M.gilvum_PYR-GCK YAVALHGKPSSTAAQQDWALSAVRKPEPDSGRFDRVW-EQVRGYDGVYTM
Mvan_0281|M.vanbaalenii_PYR-1 YAVALHGKPSSTLEQQEWALSAVRKPTVDAVRFDRVW-DQVRSYDGAYEM
MAB_3481|M.abscessus_ATCC_1997 YAVDLLGKREATASQRQWAKAVISEPVFNSERFDEIW-NRVEALSGAYEM
MMAR_0488|M.marinum_M YAVELNDKAATTDQQAQFAMRMIRRPAHDAARYDQVWKEHVLPLNGAYEM
MUL_1151|M.ulcerans_Agy99 YAVELNDKAATTDQQAQFAMRMIRRPAHDAARYDQVWKEHVLPLNGAYEM
Mb0236|M.bovis_AF2122/97 HAVELHGRSATTAEQARFAMRMVRRPVHDPARYDQIWKDHVLALNGAYQM
Rv0231|M.tuberculosis_H37Rv HAVELHGRSATTAEQARFAMRMVRRPVHDPARYDQIWKDHVLALNGAYQM
MAV_4939|M.avium_104 YAVELSDKPATTEQQARFAMRMIRRPVHDPARYDQIWKEQVLPLSGAYQM
MSMEG_1821|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
Mflv_0398|M.gilvum_PYR-GCK RP
Mvan_0281|M.vanbaalenii_PYR-1 RP
MAB_3481|M.abscessus_ATCC_1997 RA
MMAR_0488|M.marinum_M RP
MUL_1151|M.ulcerans_Agy99 RP
Mb0236|M.bovis_AF2122/97 AP
Rv0231|M.tuberculosis_H37Rv AP
MAV_4939|M.avium_104 RP
MSMEG_1821|M.smegmatis_MC2_155 --
TH_4603|M.thermoresistible__bu --