For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAAWGGNPSFDLFQLPEEHQELRAAIRALAEKEIAPHAADVDENARFPQEALDALNASGFNAVHVPEEYG GQGADSVAACIVIEEVARVDASASLIPAVNKLGTMGLILRGSDELKKQVLPQLASGAAMASYALSEREAG SDAASMRTRARADGDDWVLNGTKCWITNGGKSSWYTVMAVTDPDKGANGISAFMVHQDDEGFSVGPKERK LGIKGSPTTELYFENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIAYTKDRKQF GTSISDFQAVQFMLADMAMKVESARLMVYHAAARAERGEKNLGFISAASKCLASDIAMEVTTDAVQLFGG AGYTQDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLR
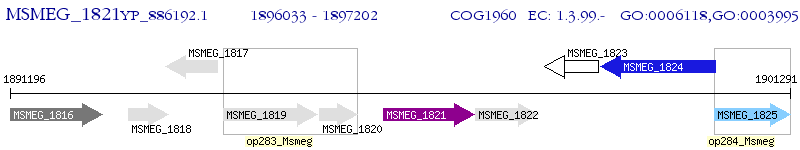
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1821 | - | - | 100% (389) | acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3982 | - | 3e-95 | 46.88% (369) | acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0108 | - | 3e-83 | 45.41% (381) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3302c | fadE25 | 0.0 | 90.49% (389) | acyl-CoA dehydrogenase FADE25 |
| M. gilvum PYR-GCK | Mflv_4743 | - | 0.0 | 93.57% (389) | butyryl-CoA dehydrogenase |
| M. tuberculosis H37Rv | Rv3274c | fadE25 | 0.0 | 90.49% (389) | acyl-CoA dehydrogenase FADE25 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 0.0 | 89.72% (389) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3618c | - | 0.0 | 90.60% (383) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_1262 | fadE25 | 0.0 | 94.86% (389) | acyl-CoA dehydrogenase FadE25 |
| M. avium 104 | MAV_4243 | - | 0.0 | 93.06% (389) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3846 | - | 0.0 | 89.97% (389) | PUTATIVE acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_2626 | fadE25 | 0.0 | 94.34% (389) | acyl-CoA dehydrogenase FadE25 |
| M. vanbaalenii PYR-1 | Mvan_1719 | - | 0.0 | 93.06% (389) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1262|M.marinum_M ----------------------------MAGWAGNPSFDLFQLPEEHQEL
MUL_2626|M.ulcerans_Agy99 ----------------------------MAGWAGNPSFDLFQLPEEHQEL
Mflv_4743|M.gilvum_PYR-GCK MQAEKSVPRNSLPAATALKLPTSSKEMLMASWAGNPSFDLFQLPEEHQEL
Mvan_1719|M.vanbaalenii_PYR-1 ----------------------------MASWAGNPSFDLFQLPEEHQEL
MSMEG_1821|M.smegmatis_MC2_155 ----------------------------MAAWGGNPSFDLFQLPEEHQEL
MAV_4243|M.avium_104 ----------------------------MAGWAGNPDFDLFQLPEEHQEL
MAB_3618c|M.abscessus_ATCC_199 -------------------------------MAGNPSFDLFKLGEEHDEL
Mb3302c|M.bovis_AF2122/97 ----------------------------MVGWAGNPSFDLFKLPEEHDEM
Rv3274c|M.tuberculosis_H37Rv ----------------------------MVGWAGNPSFDLFKLPEEHDEM
MLBr_00737|M.leprae_Br4923 ----------------------------MVGWSGNPLFDLFKLPEEHNEL
TH_3846|M.thermoresistible__bu ----------------------------MAAWSGNPSFELFQLPEEHIAL
.*** *:**:* *** :
MMAR_1262|M.marinum_M RAAIRALAEKEIAPHAADVDENSRFPQEALDALNASGFNAVHVPEEYGGQ
MUL_2626|M.ulcerans_Agy99 RAAIRALAEKEIAPHAADVDENSRFPQEALDALNASGFNALHVPEEYGGQ
Mflv_4743|M.gilvum_PYR-GCK RAAIRALAEKEIAPHAADVDENARFPQEALDALVANGFNAVHVPEEYGGQ
Mvan_1719|M.vanbaalenii_PYR-1 RAAIRALAEKEIAPHAADVDENSRFPQEALDALNASGFNAVHVPEEFGGQ
MSMEG_1821|M.smegmatis_MC2_155 RAAIRALAEKEIAPHAADVDENARFPQEALDALNASGFNAVHVPEEYGGQ
MAV_4243|M.avium_104 RAAIRALAEKEIAPHAADVDENSRFPQEALEALNAAGFNAVHVPEEYGGQ
MAB_3618c|M.abscessus_ATCC_199 RAAIRGLAEKEIAPHAKDVDEKARFPQEALDALVASGFNAVHVPEEYDGQ
Mb3302c|M.bovis_AF2122/97 RSAIRALAEKEIAPHAAEVDEKARFPEEALVALNSSGFNAVHIPEEYGGQ
Rv3274c|M.tuberculosis_H37Rv RSAIRALAEKEIAPHAAEVDEKARFPEEALVALNSSGFNAVHIPEEYGGQ
MLBr_00737|M.leprae_Br4923 RATIRALAEKEIAPHAADVDQRARFPEEALAALNASGFNAIHVPEEYGGQ
TH_3846|M.thermoresistible__bu REAIRALAEKEIAPYAAEVDEKARFPEEALAALNSSGFSAIHVPEEYGGQ
* :**.********:* :**:.:***:*** ** : **.*:*:***:.**
MMAR_1262|M.marinum_M GADSVAACIVIEEVARVDASASLIPAVNKLGTMGLILRGSDDLKKQVLPS
MUL_2626|M.ulcerans_Agy99 GADSVAACIVIEEVARVDASASLIPAVNKLGTMGLILRGSDELKKQVLPS
Mflv_4743|M.gilvum_PYR-GCK GADSVAACIVIEEVARVDASASLIPAVNKLGTMGLILRGSDELKQKVLPS
Mvan_1719|M.vanbaalenii_PYR-1 GADSVAACIVIEEVARVDASASLIPAVNKLGTMGLILRGSDELKKKVLPS
MSMEG_1821|M.smegmatis_MC2_155 GADSVAACIVIEEVARVDASASLIPAVNKLGTMGLILRGSDELKKQVLPQ
MAV_4243|M.avium_104 GADSVAACIVIEEVARVDTSASLIPAVNKLGTMGLILRGSDELKKQVLPS
MAB_3618c|M.abscessus_ATCC_199 GADSVAACIVIEEVARVCASSSLIPAVNKLGTMGLILNGSDELKKQVLPA
Mb3302c|M.bovis_AF2122/97 GADSVATCIVIEEVARVDASASLIPAVNKLGTMGLILRGSEELKKQVLPA
Rv3274c|M.tuberculosis_H37Rv GADSVATCIVIEEVARVDASASLIPAVNKLGTMGLILRGSEELKKQVLPA
MLBr_00737|M.leprae_Br4923 GADSVAACIVIEEVARVDASASLIPAVNKLGTMGLILRGSEELKKQVLPS
TH_3846|M.thermoresistible__bu GADSVATCIVIEEVARVDCSASLIPAVNKLGTMGLILRGSEELKKQVLPA
******:********** *:****************.**::**::***
MMAR_1262|M.marinum_M LADGAAMASYALSEREAGSDAGAMRTRAKADGDDWILNGTKCWITNGGKS
MUL_2626|M.ulcerans_Agy99 LADGAAMASYALSEREAGSDAGAMRTRAKADGDDWILNGTKCWITNGGKS
Mflv_4743|M.gilvum_PYR-GCK LAAGESMASYALSEREAGSDAASMRTRAKADGDDWILNGAKCWITNGGKS
Mvan_1719|M.vanbaalenii_PYR-1 IASGEAMASYALSEREAGSDAAAMRTRAKADGDDWILNGAKCWITNGGKS
MSMEG_1821|M.smegmatis_MC2_155 LASGAAMASYALSEREAGSDAASMRTRARADGDDWVLNGTKCWITNGGKS
MAV_4243|M.avium_104 IADGSAMASYALSEREAGSDAASMRTRARADGDDWILNGAKCWITNGGKS
MAB_3618c|M.abscessus_ATCC_199 IASGEAMASYALSEREAGSDAASMRTRAKADGDDWIINGSKCWITNGGKS
Mb3302c|M.bovis_AF2122/97 LAAEGAMASYALSEREAGSDAASMRTRAKADGDHWILNGAKCWITNGGKS
Rv3274c|M.tuberculosis_H37Rv LAAEGAMASYALSEREAGSDAASMRTRAKADGDHWILNGAKCWITNGGKS
MLBr_00737|M.leprae_Br4923 LAAEGAMASYALSEREAGSDAASMRTRAKADGDDWILNGFKCWITNGGKS
TH_3846|M.thermoresistible__bu VASGEAMASYALSEREAGSDAASMRTRAVADGDDWILNGSKCWITNGGKS
:* :***************.:***** ****.*::** **********
MMAR_1262|M.marinum_M SWYTVMAVTDPEKGANGISAFMVHKDDEGFVVGPKERKLGIKGSPTTELY
MUL_2626|M.ulcerans_Agy99 SWYTVMAVTDPENGANGISAFVVHKDDEGFVVGPKERKLGIKGSPTTELY
Mflv_4743|M.gilvum_PYR-GCK DWYTVMAVNDPDKGANGISAFMVHVDDEGFSIGPKERKLGIKGSPTTELY
Mvan_1719|M.vanbaalenii_PYR-1 SWYTVMAVTDPDKGANGISAFMVHIDDEGFTIGPKERKLGIKGSPTTELY
MSMEG_1821|M.smegmatis_MC2_155 SWYTVMAVTDPDKGANGISAFMVHQDDEGFSVGPKERKLGIKGSPTTELY
MAV_4243|M.avium_104 SWYTVMAVTDPDKGANGISSFMVHKDDEGFTVGPKEKKLGIKGSPTTELY
MAB_3618c|M.abscessus_ATCC_199 SWYTVMAVTDPEKKANGISAFMVHKDDEGFTVGPLEHKLGIKGSPTAELY
Mb3302c|M.bovis_AF2122/97 TWYTVMAVTDPDRGANGISAFMVHKDDEGFTVGPKERKLGIKGSPTTELY
Rv3274c|M.tuberculosis_H37Rv TWYTVMAVTDPDRGANGISAFMVHKDDEGFTVGPKERKLGIKGSPTTELY
MLBr_00737|M.leprae_Br4923 TWYTVMAVTDPDKGANGISAFIVHKDDEGFSIGPKEKKLGIKGSPTTELY
TH_3846|M.thermoresistible__bu TWYTVMAVTDPDKGANGISAFMVHKDDEGFTVGPKERKLGIKGSPTTELY
*******.**:. *****:*:** ***** :** *:*********:***
MMAR_1262|M.marinum_M FENCRVPGDRMIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIA
MUL_2626|M.ulcerans_Agy99 FENCRVPGDRMIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIA
Mflv_4743|M.gilvum_PYR-GCK FENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIE
Mvan_1719|M.vanbaalenii_PYR-1 FENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDASLE
MSMEG_1821|M.smegmatis_MC2_155 FENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIA
MAV_4243|M.avium_104 FENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIA
MAB_3618c|M.abscessus_ATCC_199 FENCRIPGDRIIGEPGTGFKTALQTLDHTRPTIGAQALGIAQGALDQAIA
Mb3302c|M.bovis_AF2122/97 FENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIA
Rv3274c|M.tuberculosis_H37Rv FENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIA
MLBr_00737|M.leprae_Br4923 FDKCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIV
TH_3846|M.thermoresistible__bu FENCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAAIA
*::**:****:************ *************:******** ::
MMAR_1262|M.marinum_M YTKDRKQFGKSISDFQAVQFMLADMAMKVESARLMVYSAAARAERGESNL
MUL_2626|M.ulcerans_Agy99 YTKDRKQFGKSISDFQAVQFMLADMAMKVESARLMVYSAAARAERGESNL
Mflv_4743|M.gilvum_PYR-GCK YTKDRKQFGKSISDFQAVQFMLADMAMKVEAARLMVYSAAARAERGEGNL
Mvan_1719|M.vanbaalenii_PYR-1 YVKDRKQFGKSISEFQAVQFMLADMAMKVEAARLMVYSAAARAERGEGNL
MSMEG_1821|M.smegmatis_MC2_155 YTKDRKQFGTSISDFQAVQFMLADMAMKVESARLMVYHAAARAERGEKNL
MAV_4243|M.avium_104 YTKERKQFGRPVSDNQGVQFMLADMAMKVESARLMVYHAAARAERGESHL
MAB_3618c|M.abscessus_ATCC_199 YTKDRKQFGKSISDFQGVQFMLADMAMKVEAARLMVYTAAARAERGEKNL
Mb3302c|M.bovis_AF2122/97 YTKDRKQFGESISTFQAVQFMLADMAMKVEAARLMVYSAAARAERGEPDL
Rv3274c|M.tuberculosis_H37Rv YTKDRKQFGESISTFQAVQFMLADMAMKVEAARLMVYSAAARAERGEPDL
MLBr_00737|M.leprae_Br4923 YTKDRKQFGESISTFQSIQFMLADMAMKVEAARLIVYAAAARAERGEPDL
TH_3846|M.thermoresistible__bu YTKERKQFGRPVSDNQGVQFMLADMAMKIEAARLMVYSAAARAERGEGDL
*.*:***** .:* *.:**********:*:***:** ********* .*
MMAR_1262|M.marinum_M GFISAASKCLASDVAMEVTTDAVQLFGGAGYTVDFPVERMMRDAKITQIY
MUL_2626|M.ulcerans_Agy99 GFISAASKCLASDVAMEVTTDAVQLFGGAGYTVDFPVERMMRDAKITQIY
Mflv_4743|M.gilvum_PYR-GCK GFISAASKCLASDVAMEVTTDAVQLFGGAGYTVDFPVERMMRDAKITQIY
Mvan_1719|M.vanbaalenii_PYR-1 GFISAASKCLASDVAMEVTTDAVQLFGGAGYTVDFPVERMMRDAKITQIY
MSMEG_1821|M.smegmatis_MC2_155 GFISAASKCLASDIAMEVTTDAVQLFGGAGYTQDFPVERMMRDAKITQIY
MAV_4243|M.avium_104 GFISAASKCLASDVAMEVTTDAVQLFGGYGYTQDFPVERMMRDAKITQIY
MAB_3618c|M.abscessus_ATCC_199 GFISSASKCFASDVAMEVTTDAVQLFGGAGYTTDFPVERMMRDAKITQIY
Mb3302c|M.bovis_AF2122/97 GFISAASKCFASDVAMEVTTDAVQLFGGAGYTTDFPVERFMRDAKITQIY
Rv3274c|M.tuberculosis_H37Rv GFISAASKCFASDVAMEVTTDAVQLFGGAGYTTDFPVERFMRDAKITQIY
MLBr_00737|M.leprae_Br4923 GFISAASKCFASDIAMEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIY
TH_3846|M.thermoresistible__bu GFISAASKCFASDVAMEVTTDAVQLFGGYGYTQDFPVERMMRDAKITQIY
****:****:***:************** *** ******:**********
MMAR_1262|M.marinum_M EGTNQIQRVVMSRALLR-
MUL_2626|M.ulcerans_Agy99 EGTNQIQRVVMSRALLR-
Mflv_4743|M.gilvum_PYR-GCK EGTNQIQRVVMSRALLK-
Mvan_1719|M.vanbaalenii_PYR-1 EGTNQIQRVVMSRALLK-
MSMEG_1821|M.smegmatis_MC2_155 EGTNQIQRVVMSRALLR-
MAV_4243|M.avium_104 EGTNQIQRVVMSRALLR-
MAB_3618c|M.abscessus_ATCC_199 EGTNQIQRVVMSRALLTS
Mb3302c|M.bovis_AF2122/97 EGTNQIQRVVMSRALLR-
Rv3274c|M.tuberculosis_H37Rv EGTNQIQRVVMSRALLR-
MLBr_00737|M.leprae_Br4923 EGTNQIQRVVMSRALLR-
TH_3846|M.thermoresistible__bu EGTNQIQRVVMSRALLR-
****************