For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSQDTNAAPKTELLHPTADDFAEFDGETRRLLKATVDWFEERGKTRLLQDYVDKVFYQDFLDFAGREKLF ATFLTPAADGAGNTEKRWDTARVAKLSEILGFYGLNYWYPWQVTVLGLGPVWQSANTDPRKRAADLLDGG AVAAFGLSEQAHGADIYSSDLVLTPVDSSEGGGYKASGRKYYIGNGNCARIVSVFGRVQGRDGLDQYVFF LADSEHPNYRVIKNVVPSQMYVAEFELDEYPVSEDDILHVGADAFSAALNTVNIGKFNLCFGGIGLSTHA FYESITHAHNRILYGNPVTKFDHVRREFVDAYARLLAMKLFSERAVDYFRTASADDRRYLLFNPVTKMKA TTEAEKVLALLADVMSAKAFEAESYTTAARIDVAGLPKLEGTVAVNLALILKFMPAYLHDAQDLPVPGVQ LQPGDDEFLFDQGPASGLGKVRFHDWRKPFADNADIPNVAAFTQRAEALATLVANERDAIKNAGLDLQLS IGELFTLVVYGGLILEQAQLRGTDSALIDQIFEFLNRDFSVYAVDLLGKREATASQRQWAKAVISEPVFN SERFDEIWNRVEALSGAYEMRA
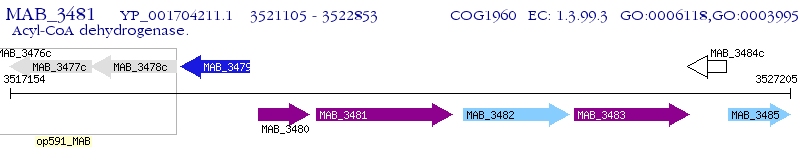
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3481 | - | - | 100% (582) | acyl-CoA dehydrogenase FadE |
| M. abscessus ATCC 19977 | MAB_3578c | - | 3e-16 | 22.92% (384) | acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3577c | - | 2e-13 | 26.32% (209) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0236 | fadE4 | 1e-166 | 52.77% (559) | acyl-CoA dehydrogenase FADE4 |
| M. gilvum PYR-GCK | Mflv_0398 | - | 0.0 | 57.17% (572) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv0231 | fadE4 | 1e-166 | 52.77% (559) | acyl-CoA dehydrogenase FADE4 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 2e-05 | 21.11% (199) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_0488 | fadE4 | 1e-160 | 50.26% (573) | acyl-CoA dehydrogenase FadE4 |
| M. avium 104 | MAV_4939 | - | 1e-166 | 53.04% (560) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0108 | - | 2e-07 | 24.34% (267) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4603 | - | 1e-07 | 23.19% (276) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_1151 | fadE4 | 1e-161 | 50.09% (573) | acyl-CoA dehydrogenase FadE4 |
| M. vanbaalenii PYR-1 | Mvan_0281 | - | 0.0 | 55.75% (583) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0398|M.gilvum_PYR-GCK ------MAARGLLLNPNHFDPAWLDPESRRLLRALIDWFEQRGKARLLRD
Mvan_0281|M.vanbaalenii_PYR-1 ------MPETPLLLDPNNFDPKRLDPESRRLLAALIDWFEQRGKVRLLRD
MAB_3481|M.abscessus_ATCC_1997 MSQDTNAAPKTELLHPTADDFAEFDGETRRLLKATVDWFEERGKTRLLQD
Mb0236|M.bovis_AF2122/97 -----------MLLNPNHLTRKYPDRRSGEIMAATVDFFESRGKARLKHD
Rv0231|M.tuberculosis_H37Rv -----------MLLNPNHLTRKYPDRRSGEIMAATVDFFESRGKARLKHD
MAV_4939|M.avium_104 -----------MLLNPNHLQRRYPDRRSAEIMAATVDFFESRGKARLKSD
MMAR_0488|M.marinum_M -----------MLLNPNRLQRKYPDARSAEIMAATVDFFESRGKARLKRD
MUL_1151|M.ulcerans_Agy99 -----------MLLNPNRLQRKYPDARSAEIMAATVDFFESRGKARLKRD
MSMEG_0108|M.smegmatis_MC2_155 ---------MS-------VDRLLPSEEARDLIALTREVADKVLDPIVDEH
TH_4603|M.thermoresistible__bu ---------MA-------VDRLLPSQEAAELIELTREIADKVLDPIVDRH
MLBr_00737|M.leprae_Br4923 ---------MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADV
. : :.
Mflv_0398|M.gilvum_PYR-GCK DLDGVWVADFLEFVKKERLFATFLTPSEFAEGDPNKRWDTARNAVLSEIL
Mvan_0281|M.vanbaalenii_PYR-1 DLNAEWVTDFLDFVRRERLFATFLTPSEFANGDPNKRWDTSRNAVLSEIL
MAB_3481|M.abscessus_ATCC_1997 YVDKVFYQDFLDFAGREKLFATFLTPAADGAGNTEKRWDTARVAKLSEIL
Mb0236|M.bovis_AF2122/97 DHERIWYSDFLDFVGRERIFASLLTPASYGADD--CRWDTYRISEFAEIM
Rv0231|M.tuberculosis_H37Rv DHERIWYSDFLDFVGRERIFASLLTPASYGADD--CRWDTYRISEFAEIM
MAV_4939|M.avium_104 DHERVWYSDFLEHIGRERIFASLLTPTRYGSAD--CRWDTYRISGFAEIL
MMAR_0488|M.marinum_M DHDRAWYSDFLNHIAEHRIFASLLTPSEYGADD--CRWDTYRISEFAEII
MUL_1151|M.ulcerans_Agy99 DHDRAWYSDFLNHIAEHRIFASLLTPSEYGADD--CRWDTYRISEFAEII
MSMEG_0108|M.smegmatis_MC2_155 ERNETYPEGVFAQLG-AAGLLSLPQPEQWGGGG---QPYEVYLQVLEEIA
TH_4603|M.thermoresistible__bu EKDETYPEGVFEQLG-AAGLLSLPQPEEWGGGG---QPYEVYLQVLEEIA
MLBr_00737|M.leprae_Br4923 DQRARFPEEALAALN-ASGFNAIHVPEEYGGQG---ADSVAACIVIEEVA
: : : :: * . . : *:
Mflv_0398|M.gilvum_PYR-GCK GFYGLSYWYAEQVTILGLGPIWQSDNVKARERAAAQLEAGEVMAFGLSER
Mvan_0281|M.vanbaalenii_PYR-1 GFYGLSYWYAEQVTILGLGPIWQSGNVKAKERAAEQLEAGGVMAFGLSER
MAB_3481|M.abscessus_ATCC_1997 GFYGLNYWYPWQVTVLGLGPVWQSANTDPRKRAADLLDGGAVAAFGLSEQ
Mb0236|M.bovis_AF2122/97 GFYGLSYWYPFQVTALGLGPIWMSANEDAKRKAAAGLEAGEVFAFGLSEQ
Rv0231|M.tuberculosis_H37Rv GFYGLSYWYPFQVTALGLGPIWMSANEDAKRKAAAGLEAGEVFAFGLSEQ
MAV_4939|M.avium_104 GFYGLNYWYPFQVTALGLGPIWMSGNDEAKRKAAGLLEAGEVFAFGLSEQ
MMAR_0488|M.marinum_M GFYGLSYWYPFQVTALGLGPIWMSDNEDAKRKAAAQLEQGEVFAFGLSEQ
MUL_1151|M.ulcerans_Agy99 GFYGLSYWYPFQVTALGLGPIWMSDNEDAKRKAAAQLEQGEVFAFGLSEQ
MSMEG_0108|M.smegmatis_MC2_155 ARWAAVAVAVSVHSLSSHPLLAFGTDEQKQRWLPGMLSGEQIGAYSLSEP
TH_4603|M.thermoresistible__bu ARWASVAVAVSVHSLSSHPLLVFGTEEQKKRWLPGMLSGEQIGAYSLSEP
MLBr_00737|M.leprae_Br4923 RVDASASLIPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSER
. . : . : :. . : ::.***
Mflv_0398|M.gilvum_PYR-GCK AHGADIYNTDMLLTPASP-----------GADDPVVFRASGEKYYIGNGN
Mvan_0281|M.vanbaalenii_PYR-1 DHGADIYNTDMLLTPAGDERSREEQTTADGDDDGVLFRASGEKYYIGNGN
MAB_3481|M.abscessus_ATCC_1997 AHGADIYSSDLVLTPVDS-------------SEGGGYKASGRKYYIGNGN
Mb0236|M.bovis_AF2122/97 THGADVYQTDMILTPS-----------------DGGWTANGEKYYIGNAN
Rv0231|M.tuberculosis_H37Rv THGADVYQTDMILTPS-----------------DGGWTANGEKYYIGNAN
MAV_4939|M.avium_104 THGADVYQTDMNLTPAP----------------GGGWTAAGQKYYIGNAN
MMAR_0488|M.marinum_M AHGADVYQTDMILTPS-----------------EHGWTASGEKYYIGNAN
MUL_1151|M.ulcerans_Agy99 AHGADVYQTDMILTPS-----------------EHGWTASGEKYYIGNAN
MSMEG_0108|M.smegmatis_MC2_155 QAGSDAAALRCAARRD-----------------GDGFVLNGSKAWITHGG
TH_4603|M.thermoresistible__bu QAGSDAAALRCAATPT-----------------DGGYVINGSKSWITHGG
MLBr_00737|M.leprae_Br4923 EAGSDAASMRTRAKAD-----------------GDDWILNGFKCWITNGG
*:* : * * :* :..
Mflv_0398|M.gilvum_PYR-GCK VAGMVSVFGRRTDVEGADGYVWFVADSRHPDYELIGNVVHG---QMFVSN
Mvan_0281|M.vanbaalenii_PYR-1 VAGMVSVFARRTDVEGADGYVWFVADSGHERYELIGNVVHG---QMFVSN
MAB_3481|M.abscessus_ATCC_1997 CARIVSVFGRVQGRDGLDQYVFFLADSEHPNYRVIKNVVPS---QMYVAE
Mb0236|M.bovis_AF2122/97 VARMVSTFGKIAGTPESQEYVFFVADSQHERYDLIKNVVNS---QNYVAN
Rv0231|M.tuberculosis_H37Rv VARMVSTFGKIAGTPESQEYVFFVADSQHERYDLIKNVVNS---QNYVAN
MAV_4939|M.avium_104 VARMVSTFGKFAGGATDSEYVFFAADSRHERYRLIKNVVNS---QNYVGN
MMAR_0488|M.marinum_M VARMVSTFGKIEDSSKEPEYVFFAADSQHDRYELKKNVVNS---QNYVAN
MUL_1151|M.ulcerans_Agy99 VARMVSTFGKIEDSSKEPEYVFFAADSQHDRYELKKNVVNS---QNYVAN
MSMEG_0108|M.smegmatis_MC2_155 RADFYTLFARTG--EGSKGVSCFLVPGDLPGLSFGKPEEKMGLHAVPTTS
TH_4603|M.thermoresistible__bu KADFYTLFARTG--EGSRGVSCFLVPADQPGLSFGKPEEKMGLHAVPTTS
MLBr_00737|M.leprae_Br4923 KSTWYTVMAVTDPDKGANGISAFIVHKDDEGFSIGPKEKKLGIKGSPTTE
: : :. * . . . .
Mflv_0398|M.gilvum_PYR-GCK FALRDYPVYEEDILATGQDAFSAALNTVNVGKFNLCSGSIGMCEHAFYEA
Mvan_0281|M.vanbaalenii_PYR-1 FALHDYPVHEEDILATGPDAFSAALNTVNVGKFNLCSGSIGMCEHAFYEA
MAB_3481|M.abscessus_ATCC_1997 FELDEYPVSEDDILHVGADAFSAALNTVNIGKFNLCFGGIGLSTHAFYES
Mb0236|M.bovis_AF2122/97 YALRDYPVTEADILHRGAEAFHAALNTVNVCKYNLGWGAIGMCTHALYES
Rv0231|M.tuberculosis_H37Rv YALRDYPVTEADILHRGAEAFHAALNTVNVCKYNLGWGAIGMCTHALYES
MAV_4939|M.avium_104 FALSDYPVTEADILHRGPEAFHAALNTVNVCKYNLGWGAVGMCTHAMYEA
MMAR_0488|M.marinum_M YALHDYPVTEADLLHRGMGAFHAALNTVNVCKYNLGWGAVGMCTHAFYEA
MUL_1151|M.ulcerans_Agy99 YALHDYPVTEADLLHRGMGAFHAALNTVNVCKYNLGWGAVGMCTHAFYEA
MSMEG_0108|M.smegmatis_MC2_155 AFYDNARIDADRLIGTEGQGLSIAFSALDSGRLGIAAVAVGIAQAALDEA
TH_4603|M.thermoresistible__bu AFYDNARIDADRRIGEEGQGLQIAFSALDSGRLGIAAVATGLAQAALDEA
MLBr_00737|M.leprae_Br4923 LYFDKCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALDAA
. : : .: *: ::: : : . *:. *: :
Mflv_0398|M.gilvum_PYR-GCK ITHANNRILYGNPVTDFPHVRATFVDAYARLIAMKLFSDRAIDYFRSAGP
Mvan_0281|M.vanbaalenii_PYR-1 INHANNRILYGNPVTAFPHVRAGFVDAYTRLIAMKLFSDRAIDYFRSAGP
MAB_3481|M.abscessus_ATCC_1997 ITHAHNRILYGNPVTKFDHVRREFVDAYARLLAMKLFSERAVDYFRTASA
Mb0236|M.bovis_AF2122/97 VTHAANRHLYGTVVTDFSHVRRLLTDAYVRLIAMKLVASRASDYMRSASA
Rv0231|M.tuberculosis_H37Rv VTHAANRHLYGTVVTDFSHVRRLLTDAYVRLIAMKLVASRASDYMRSASA
MAV_4939|M.avium_104 VTHASNRILYGTVVTDFSHVRRLLIDAYARLVAMRLVATRACDYMRSASA
MMAR_0488|M.marinum_M ITHAANRHLYGTVVTDFSHVRRLLTDAYARLVAMRLVASRASDYMRSASA
MUL_1151|M.ulcerans_Agy99 ITHAANRHLYGTVVTDFSHVRRLLTDAYARLVAMRLVASRASDYMRSASA
MSMEG_0108|M.smegmatis_MC2_155 VRYAHERTTFGRKIIDHQGLGFLLADMAAAVVSARATYLDAARRRDAGLP
TH_4603|M.thermoresistible__bu VAYANERTAFGRKIIDHQGLGFLLADMAAAVATARATYLDAARRRDQGRP
MLBr_00737|M.leprae_Br4923 IVYTKDRKQFGESISTFQSIQFMLADMAMKVEAARLIVYAAAARAERGEP
: :: :* :* : . : : * : : : * . .
Mflv_0398|M.gilvum_PYR-GCK HDRRYLLFNPVTKSKVTSEGEKVVTLLWDVLAAKGFEKDTYFYQVTRLIG
Mvan_0281|M.vanbaalenii_PYR-1 DDRRYLLFNPVTKSKVTSEGEKVVTLLWDVLAAKGFEKNTYFYQVTRLIG
MAB_3481|M.abscessus_ATCC_1997 DDRRYLLFNPVTKMKATTEAEKVLALLADVMSAKAFEAESYTTAARIDVA
Mb0236|M.bovis_AF2122/97 ADRRYLLYSPLTKAKVTSEGERVITALWDVIAAKGVEKDTFFETVAREIG
Rv0231|M.tuberculosis_H37Rv ADRRYLLYSPLTKAKVTSEGERVITALWDVIAAKGVEKDTFFETVAREIG
MAV_4939|M.avium_104 TDRRYLLYSPLTKAKITGEGERVITALWDVIAAKGVEKDTIFEAMTREIG
MMAR_0488|M.marinum_M EDRRYLLYSPLTKAKVTSEGERVIAALWDVIAAKGVEKDTFFETVAREIG
MUL_1151|M.ulcerans_Agy99 EDRRYLLYSPLTKAKVTSEGERVIAALWDVIAAKGVEKDTFFETVAREIG
MSMEG_0108|M.smegmatis_MC2_155 ----YSSQASVAKLVATDAAMKVTTDAVQVLGGVGYTRDFRVERYMREAK
TH_4603|M.thermoresistible__bu ----YSQQASIAKLTATDAAMKVTTDAVQVFGGVGYTRDYRVERYMREAK
MLBr_00737|M.leprae_Br4923 D---LGFISAASKCFASDIAMEVTTDAVQLFGGAGYTSDFPVERFMRDAK
. :* : . .* : :::.. . :
Mflv_0398|M.gilvum_PYR-GCK ALPRLEGTVHVNVAQILKFMPNYMFNP-AEYPPIGTRDDPADDVFFWSQG
Mvan_0281|M.vanbaalenii_PYR-1 ALPRLEGTVHVNVAQILKFMPNFMFNP-APYPEIGTRDDPADDVFFWSQG
MAB_3481|M.abscessus_ATCC_1997 GLPKLEGTVAVNLALILKFMPAYLHDA-QDLPVPGVQLQPGDDEFLFDQG
Mb0236|M.bovis_AF2122/97 LLPRLEGTVHINIGLLGKFMPNYLFAPDSTLPVIPRRDDAADDAFLFAQG
Rv0231|M.tuberculosis_H37Rv LLPRLEGTVHINIGLLGKFMPNYLFAPDSTLPVIPRRDDAADDAFLFAQG
MAV_4939|M.avium_104 LLPRLEGTVHINIGLLGKFMPNYLFAPSGELPVIGRRDDAADDEFLFAQG
MMAR_0488|M.marinum_M LLPRLEGTVHINIGLLAKFMPNFLFVPDAALPLIPRRDDAADDTFLFNQG
MUL_1151|M.ulcerans_Agy99 LLPRLEGTVHINIGLLAKFMPNFLFAPDAALPLIPRRDDAADDTFLFNQG
MSMEG_0108|M.smegmatis_MC2_155 ITQIFEGTNQIQRLVISRGLGT----------------------------
TH_4603|M.thermoresistible__bu IMQIFEGTNQIQRLVIARGLTR----------------------------
MLBr_00737|M.leprae_Br4923 ITQIYEGTNQIQRVVMSRALLR----------------------------
*** :: : : :
Mflv_0398|M.gilvum_PYR-GCK PARGASKVRFADWAPVYERHAGIPNVGRFYEQVQAFKQLLTVSAPDADQQ
Mvan_0281|M.vanbaalenii_PYR-1 PARGASKVRFADWAPVYERNTGIPNVAVFYQQVQAFKDLLATAAPDADQQ
MAB_3481|M.abscessus_ATCC_1997 PASGLGKVRFHDWRKPFADNADIPNVAAFTQRAEALATLVANER-DAIKN
Mb0236|M.bovis_AF2122/97 PTGGLGKVRFHDWRASFDTCAHLPNVALLREQVDVFAELLASATPDAAQQ
Rv0231|M.tuberculosis_H37Rv PTGGLGKVRFHDWRASFDTCAHLPNVALLREQVDVFAELLASATPDAAQQ
MAV_4939|M.avium_104 PTGGLGKVRFGDWRAPFDGFAHLPNVALLRAQIDVLAEMLASATPDAVQQ
MMAR_0488|M.marinum_M PTGGLGKVRFHDWRAPFDSYGHLPNVALLREQIDVLTEMLAGATPDASQQ
MUL_1151|M.ulcerans_Agy99 PTGGLGKVRFHDWRAPFGSYGHLPNVALLREQIDVLTEMLAGATPDASQQ
MSMEG_0108|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0398|M.gilvum_PYR-GCK ADLDFVLVVGHLFSLVVYGQLILEQAEITGLDRDVLDQIFDVAVRDFSAY
Mvan_0281|M.vanbaalenii_PYR-1 RDLDFVLVVGHLFTLVVYGQLILEQAELTGLESDLVDQIFDVQIRDFSAY
MAB_3481|M.abscessus_ATCC_1997 AGLDLQLSIGELFTLVVYGGLILEQAQLRGTDSALIDQIFEFLNRDFSVY
Mb0236|M.bovis_AF2122/97 KDIDFAFGVGQLFANVPYAQLILEEARLSGVDEALIDEIFGVLVRDFNTH
Rv0231|M.tuberculosis_H37Rv KDIDFAFGVGQLFANVPYAQLILEEARLSGVDEALIDEIFGVLVRDFNTH
MAV_4939|M.avium_104 KDIDFAFAVGQLFATVPYAQLILEEATLSGVDDALIDEIFGVLVRDFNAY
MMAR_0488|M.marinum_M KDIDFAFGVGQIFALVPYAQLILEEAPLSGVDDALLDEIFGLLVRDFNSY
MUL_1151|M.ulcerans_Agy99 KDIDFAFGVGQIFALVPYAQLILEEAPLSGVDDALLDEIFGLLVRDFNSY
MSMEG_0108|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0398|M.gilvum_PYR-GCK AVALHGKPSSTAAQQDWALSAVRKPEPDSGRFDRVW-EQVRGYDGVYTMR
Mvan_0281|M.vanbaalenii_PYR-1 AVALHGKPSSTLEQQEWALSAVRKPTVDAVRFDRVW-DQVRSYDGAYEMR
MAB_3481|M.abscessus_ATCC_1997 AVDLLGKREATASQRQWAKAVISEPVFNSERFDEIW-NRVEALSGAYEMR
Mb0236|M.bovis_AF2122/97 AVELHGRSATTAEQARFAMRMVRRPVHDPARYDQIWKDHVLALNGAYQMA
Rv0231|M.tuberculosis_H37Rv AVELHGRSATTAEQARFAMRMVRRPVHDPARYDQIWKDHVLALNGAYQMA
MAV_4939|M.avium_104 AVELSDKPATTEQQARFAMRMIRRPVHDPARYDQIWKEQVLPLSGAYQMR
MMAR_0488|M.marinum_M AVELNDKAATTDQQAQFAMRMIRRPAHDAARYDQVWKEHVLPLNGAYEMR
MUL_1151|M.ulcerans_Agy99 AVELNDKAATTDQQAQFAMRMIRRPAHDAARYDQVWKEHVLPLNGAYEMR
MSMEG_0108|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_0398|M.gilvum_PYR-GCK P
Mvan_0281|M.vanbaalenii_PYR-1 P
MAB_3481|M.abscessus_ATCC_1997 A
Mb0236|M.bovis_AF2122/97 P
Rv0231|M.tuberculosis_H37Rv P
MAV_4939|M.avium_104 P
MMAR_0488|M.marinum_M P
MUL_1151|M.ulcerans_Agy99 P
MSMEG_0108|M.smegmatis_MC2_155 -
TH_4603|M.thermoresistible__bu -
MLBr_00737|M.leprae_Br4923 -