For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSLPITLGWVFVAVALGVLSPSLDDVASQHSVPMTPRDAPAFKSMMHIGDKFNEFHTDSTAMVVLEGKDK LGDSAHEFYDRIVRKLRDDHQHVQNIQDFWSDPLTAAGSQSPDGKSAYVQVFLNGAQGTTPSYESVAAVR KIVDSTPAPPGVKAYVAGNTVLNADTSIVGHKSMATMALVSIVVIFVMLLVVYRSIVTTVLSLVIIGIEL FAAQGITATAGNLNIIGLTPYAVSMITMLSIAAGTDYVIFLLGRYHEARSFGQGREEAFYTAYHGVSHVI LGSGLTIAGACLCLTAARLPYFQTMGLPCAIAMVVIVLAALTLAPAILAVGSRFGLFDPKRAIDVRGWRK VGTAVVRWPKPIIVVTAAIAVIGFISLLTYVPNYDDQKFTPKDMPANAAMTAAERHFSQARMNPELLMVE ADHDLRDPADMLVVDRIAKSVFHLRGIERVQTITRPLGAPIEHSSIPFQIAMQNSGTLQTAKFMNDTMAN MLEQADELDKTIAVMEHMYGVMKELTATTHSMVGRTHEMVDTTNELRDRIADFDDFFRPIRSYFYWEKHC FDIPVCWSMRSLFDALDGIDELSDQISGLTIDLDHMDRLMPQLLADFPKTIDSMKTMRDFMLSTHSTMAG IQAHQQEAAEGSTLMGQYFDEAKNDDSFYLPPEVFKNPDFKRGLKMFVSPDGTAVRFIITHQGDPASVEG IKHVAGVKDAVADAIKGTPLESSKVYLAGTASMYSDMQEGVIIDLLVAGISCLILIFTIMLIITRSVVAA LVIVGTVAASLGTACGLSVLMWQDLIGLGVQWIVLPLSIVILLAVGSDYNLLLVSRLKEEIPAGLNTGII RGMGASGRVVTAAGLVFAFTMASMIVSQLRVIGELGTTIALGLLVDTLIVRSFMTPSIAAALGHWFWWPI NTIRMTRRTHHDEEAHTAPIRQPALR*
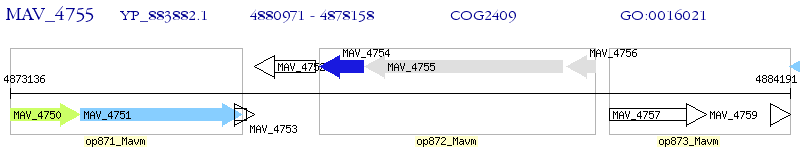
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4755 | - | - | 100% (937) | MmpL4 protein |
| M. avium 104 | MAV_4696 | - | 0.0 | 57.98% (909) | MmpL4 protein |
| M. avium 104 | MAV_2510 | - | 0.0 | 57.21% (909) | MmpL4 protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0458c | mmpL4 | 0.0 | 60.02% (918) | transmembrane transport protein MmpL4 |
| M. gilvum PYR-GCK | Mflv_4418 | - | 0.0 | 57.80% (910) | transport protein |
| M. tuberculosis H37Rv | Rv0450c | mmpL4 | 0.0 | 60.13% (918) | transmembrane transport protein MmpL4 |
| M. leprae Br4923 | MLBr_02378 | - | 0.0 | 58.04% (908) | hypothetical protein MLBr_02378 |
| M. abscessus ATCC 19977 | MAB_4310c | - | 0.0 | 72.67% (933) | MmpL family protein |
| M. marinum M | MMAR_0775 | - | 0.0 | 61.17% (909) | MmpL family transport protein |
| M. smegmatis MC2 155 | MSMEG_0463 | - | 0.0 | 74.21% (919) | MmpL4 protein |
| M. thermoresistible (build 8) | TH_3260 | - | 0.0 | 52.51% (937) | PUTATIVE MmpL4 protein |
| M. ulcerans Agy99 | MUL_0757 | mmpL5 | 0.0 | 57.10% (909) | transmembrane transport protein, MmpL5 |
| M. vanbaalenii PYR-1 | Mvan_1936 | - | 0.0 | 57.25% (910) | transport protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4418|M.gilvum_PYR-GCK ---------------------------------MADAGRSGIGRIIRVLA
Mvan_1936|M.vanbaalenii_PYR-1 ---------------------------------MADAGRSGIGRIIHLLA
TH_3260|M.thermoresistible__bu ------------MTANPGHVRTDEAPTDRLDIKVSTHGPSFISTWIRRLS
MUL_0757|M.ulcerans_Agy99 ------------MTAT-----TTDTPTDVVPPAEHPPRP-FIPRMIRTFA
MMAR_0775|M.marinum_M MSDDQAVSERAASNGASNGAGAENGNGNENGNGNGNGSRPSMARLIHRMA
Mb0458c|M.bovis_AF2122/97 ----------------------MSTKFANDSNTNARPEKPFIARMIHAFA
Rv0450c|M.tuberculosis_H37Rv ----------------------MSTKFANDSNTNARPEKPFIARMIHAFA
MLBr_02378|M.leprae_Br4923 ----------------------MTVKCANDLDTHTKP--PFVARMIHRFA
MAB_4310c|M.abscessus_ATCC_199 -----------------------------MSNLHAKPHRPFLGHLIRVLS
MSMEG_0463|M.smegmatis_MC2_155 -----------------------------MTNTHSKSQRPFVGHVIRIFS
MAV_4755|M.avium_104 -----------------------------------------------MLS
::
Mflv_4418|M.gilvum_PYR-GCK VPIVLGWVLLTVLTNVAVPSLEKVGEEHTVGMSAGDAPSMMSMKRVGANF
Mvan_1936|M.vanbaalenii_PYR-1 VPIVLGWIALTVLTNVAVPSLEKVGEEHTVGMSANDAPSMMSMERVGANF
TH_3260|M.thermoresistible__bu LPIIVGWIALIALLNITVPQLEVVGEMRAVSMSPDEAPSMVAMKRVGELF
MUL_0757|M.ulcerans_Agy99 VPLILGWVLLIAVLNSVVPQLEVVGQMQAVSMSPDAAPSMISMKHIGKVF
MMAR_0775|M.marinum_M VPIVVFWVGLVAVLVMFVPSLEEVGKQHTVSLSPSDAESMIAMKRVGKVF
Mb0458c|M.bovis_AF2122/97 VPIILGWLAVCVVVTVFVPSLEAVGQERSVSLSPKDAPSFEAMGRIGMVF
Rv0450c|M.tuberculosis_H37Rv VPIILGWLAVCVVVTVFVPSLEAVGQERSVSLSPKDAPSFEAMGRIGMVF
MLBr_02378|M.leprae_Br4923 VPIILVWLAIAVTVSVFIPSLEDVGQERSVSLSPKDAPSYIAMQKMGQVF
MAB_4310c|M.abscessus_ATCC_199 LPIILFWVALAVILGTVTPSLTDVAASRSVPLSPQNSESYQGMLNIGKVF
MSMEG_0463|M.smegmatis_MC2_155 LPIVLLWVLLVVALGAIFPSLDDVAATRSVPLSPTDSQAYQSMLNIGKVF
MAV_4755|M.avium_104 LPITLGWVFVAVALGVLSPSLDDVASQHSVPMTPRDAPAFKSMMHIGDKF
:*: : *: : . *.* *. ::* ::. : : .* .:* *
Mflv_4418|M.gilvum_PYR-GCK EEFDSDSSAMVVLEGEQPLGDAAHRYYDQLIDELEADTANVQHIADFWGD
Mvan_1936|M.vanbaalenii_PYR-1 DEFDSDSSAMVVLEGEQPLGDAAHRYYDELIGKLEADTANVQHVADFWGD
TH_3260|M.thermoresistible__bu DEGSSDSQVMIVLEGEEPLGADARAFYDELVKRLEADDAHVENVQDLWSD
MUL_0757|M.ulcerans_Agy99 REGDSDSAAMIVLEGQEPLGAAAHKFYDKMITKLRADTKHVQSVQDFWGD
MMAR_0775|M.marinum_M NEFDTDSAVMIVLEGDHPLGDEAHHYYDELVRKLKADKDHVQHVQDFWGD
Mb0458c|M.bovis_AF2122/97 KEGDSDSFAMVIIEGNQPLGDAAHKYYDGLVAQLRADKKHVQSVQDLWGD
Rv0450c|M.tuberculosis_H37Rv KEGDSDSFAMVIIEGNQPLGDAAHKYYDGLVAQLRADKKHVQSVQDLWGD
MLBr_02378|M.leprae_Br4923 NEGNSDSVIMIVLEGDKPLGDDAHRFYDGLIRKLRADK-HVQSVQDFWGD
MAB_4310c|M.abscessus_ATCC_199 QQYDSDSSAMVVLEGDDKLGDSAHQYYDDIVAKLKADHEHVQNVQDFWSD
MSMEG_0463|M.smegmatis_MC2_155 RQYDSDSTAMVVLEGEDKLGDAAHKYYDEIVAKLRADHEHVQNVQDFWSD
MAV_4755|M.avium_104 NEFHTDSTAMVVLEGKDKLGDSAHEFYDRIVRKLRDDHQHVQNIQDFWSD
: :** *:::**.. ** *: :** :: .*. * :*: : *:*.*
Mflv_4418|M.gilvum_PYR-GCK PLTASGAQSTDGKAAYVQVYLQGNQGEPRANEAVAAVREIVDRTP---AP
Mvan_1936|M.vanbaalenii_PYR-1 PLTASGAQSTDGKAAYVQVYLQGNQGEPRANEAVAAVREIVDQTP---AP
TH_3260|M.thermoresistible__bu PLTATGVQSPDGKSAYVQVKLAGNQGEALANESVETVREMIATVSEEMAP
MUL_0757|M.ulcerans_Agy99 PLTATGAQSGDGKAAYVQVKLAGNQGESLANESVEAVKTIVDSL---QAP
MMAR_0775|M.marinum_M PLTAAGSQSPDGKAAYVQAYLVGNQGEAKANESVDAVRELVNDTP---AP
Mb0458c|M.bovis_AF2122/97 PLTAAGVQSNDGKAAYVQLSLAGNQGTPLANESVEAVRSIVESTP---AP
Rv0450c|M.tuberculosis_H37Rv PLTAAGVQSNDGKAAYVQLSLAGNQGTPLANESVEAVRSIVESTP---AP
MLBr_02378|M.leprae_Br4923 PLTAPGAQSNDGKAAYVQLSLAGNQGELLAQESIDAVRKIVVQTP---AP
MAB_4310c|M.abscessus_ATCC_199 PLTAAGSQSVDGKAAYVQIFLNGSQGTSASHESVAAVRHIVDSVP---AP
MSMEG_0463|M.smegmatis_MC2_155 PLTAAGSQSVDGRSAYVQIFLNGPQGTTASYESVAAVRDIVESVP---PP
MAV_4755|M.avium_104 PLTAAGSQSPDGKSAYVQVFLNGAQGTTPSYESVAAVRKIVDSTP---AP
****.* ** **::**** * * ** : *:: :*: :: .*
Mflv_4418|M.gilvum_PYR-GCK PGIKAYVTGGAPLVADQHSAGDTSVLRVTLITIGVIAVMLLIVFRSVATM
Mvan_1936|M.vanbaalenii_PYR-1 PGLKAYVTGGAPLVADQHNAGDKSVFRVTLITIGVIAVMLLLVFRSVATM
TH_3260|M.thermoresistible__bu EGVQAFVTGGAAMSADQQEAGDSSLRVIEALTFLVITTMLLFVYRSVITV
MUL_0757|M.ulcerans_Agy99 PGVKVYVTGSAALVADQQLAGDRSLQLIEMVTFTVIIVMLLLVYRSIITA
MMAR_0775|M.marinum_M PGVKAYVTGPAALIADQSTAGDASIQRVTFITIGVIFVMLLSVYRSLITV
Mb0458c|M.bovis_AF2122/97 PGIKAYVTGPSALAADMHHSGDRSMARITMVTVAVIFIMLLLVYRSIITV
Rv0450c|M.tuberculosis_H37Rv PGIKAYVTGPSALAADMHHSGDRSMARITMVTVAVIFIMLLLVYRSIITV
MLBr_02378|M.leprae_Br4923 PGITAWVTGASALIADMHHSGDKSMIRITATSVIVILVTLLLVYRSFITV
MAB_4310c|M.abscessus_ATCC_199 PGIKAHVAGTTVLNADTQVAGHQSMATMELVSVGVIIVMILFIYRSIVTM
MSMEG_0463|M.smegmatis_MC2_155 PGIKAHVAGTTVLNADTSVAGHQSMARMELISVAVIILMLLAIYRSIVTM
MAV_4755|M.avium_104 PGVKAYVAGNTVLNADTSIVGHKSMATMALVSIVVIFVMLLVVYRSIVTT
*: . *:* : : ** *. *: : :. ** :* ::**. *
Mflv_4418|M.gilvum_PYR-GCK VLILFMVFAELGAARGIVAFLADAEIIGLSTFATSLLTLMVIAAGTDYAI
Mvan_1936|M.vanbaalenii_PYR-1 VLILFMVFAELGAARGIVAFLANAEIIGLSTFATSLLTLMVIAAGTDYAI
TH_3260|M.thermoresistible__bu LIALFMVLIGLLTTRGVVAFLGYHNIIGLSTFATSLLVTLAIAIAVDYAI
MUL_0757|M.ulcerans_Agy99 AIMLTMVVLGLLATRGGVAFLGYHRIIGLSTFATNLLVVLAIAAATDYAI
MMAR_0775|M.marinum_M ISVLVMVGIELMAARGVVAFLADNNVIGLSTFAVNLLVLMAIAAGTDYAI
Mb0458c|M.bovis_AF2122/97 VLLLITVGVELTAARGVVAVLGHSGAIGLTTFAVSLLTSLAIAAGTDYGI
Rv0450c|M.tuberculosis_H37Rv VLLLITVGVELTAARGVVAVLGHSGAIGLTTFAVSLLTSLAIAAGTDYGI
MLBr_02378|M.leprae_Br4923 ILLLFTVGIESAVARGVVALLGHTGLIGLSTFAVNLLTSLAIAAGTDYGI
MAB_4310c|M.abscessus_ATCC_199 LISMVIIGLELFAAQGVTATAGYLNVIGLTPYAVSMVTMLALAAGTDYVI
MSMEG_0463|M.smegmatis_MC2_155 LISMVIIGLELFAAQSVTATAGHFNLIGLTPYAVSMTTMLALAAGTDYVI
MAV_4755|M.avium_104 VLSLVIIGIELFAAQGITATAGNLNIIGLTPYAVSMITMLSIAAGTDYVI
: : .::. .* . ***:.:*..: . : :* ..** *
Mflv_4418|M.gilvum_PYR-GCK FAIGRYQEARGAGEDRETAYYTMFRGTAHVVLGSGLTIAGAMLCLSFTRL
Mvan_1936|M.vanbaalenii_PYR-1 FAIGRYQEARGAGEDRETAYYTMFRGTAHVVLGSGLTIAGAMLCLSFTRL
TH_3260|M.thermoresistible__bu FLIGRYQEARSVGADRATAYHTMFGGTAHVILASGLTIAGATFCLSFTRL
MUL_0757|M.ulcerans_Agy99 FLIGRYQEARGLGQDRESAYYTMFGGTAHVVLGSGLTIAGATFCLSFTRL
MMAR_0775|M.marinum_M FVLGRYQEARGEGESREKAFYTMFHGTAHVVLGSGLTIAGAMYCLSFTRL
Mb0458c|M.bovis_AF2122/97 FIIGRYQEARQAGEDKEAAYYTMYRGTAHVILGSGLTITGATFCLSFARM
Rv0450c|M.tuberculosis_H37Rv FIIGRYQEARQAGEDKEAAYYTMYRGTAHVILGSGLTIAGATFCLSFARM
MLBr_02378|M.leprae_Br4923 FITGRYQEARQANENKEAAFYTMYRGTFHVILGSGLTISGATFCLSFARM
MAB_4310c|M.abscessus_ATCC_199 FLFGRYHEERSKGLSREDAYYVAYHGVSHVILGSGLTIVGACLCLTMTTL
MSMEG_0463|M.smegmatis_MC2_155 FLLGRYHEERSRGLDREEAYYVAYRGVSHVILGSGLTIAGACLCLTMTTL
MAV_4755|M.avium_104 FLLGRYHEARSFGQGREEAFYTAYHGVSHVILGSGLTIAGACLCLTAARL
* ***:* * . .: *::. : *. **:*.***** ** **: : :
Mflv_4418|M.gilvum_PYR-GCK PYFQTMGVPCAVGTFVAVIAALTLGPAVIVIGSRFG-LFDPKRAISSRGW
Mvan_1936|M.vanbaalenii_PYR-1 PYFQTMGVPCAVGTFVAVMAALTLGPAVVAIGSRFG-LFDPKRAISSRGW
TH_3260|M.thermoresistible__bu PYFQSLGPPLAIGMIVLIVTSLTMGPAIITVATRFG-LLEPKRAARVRFW
MUL_0757|M.ulcerans_Agy99 PYFQTLGVPLAIGMVIVVATALTLGPALIAVMSRFGKLLEPKRMARVRGW
MMAR_0775|M.marinum_M PYFQTLGAPCAVGMLVAVLAALTLGPAVLVVGSFFK-LFDPKRKMRTRGW
Mb0458c|M.bovis_AF2122/97 PYFQTLGIPCAVGMLVAVAVALTLGPAVLHVGSRFG-LFDPKRLLKVRGW
Rv0450c|M.tuberculosis_H37Rv PYFQTLGIPCAVGMLVAVAVALTLGPAVLHVGSRFG-LFDPKRLLKVRGW
MLBr_02378|M.leprae_Br4923 PYFQTLGVPCAVGMLIAVAVALTLGPAVLTVGSRFG-LFEPKRLIKVRGW
MAB_4310c|M.abscessus_ATCC_199 PYFQSMALPCAISVLVIVAAALTLAPAVLTVASKFG-LLEPRGKLSTTGW
MSMEG_0463|M.smegmatis_MC2_155 PYFQSMALPCAISILVIVIAALTLAPAVLTVASRFG-LLDPKRELSTKSW
MAV_4755|M.avium_104 PYFQTMGLPCAIAMVVIVLAALTLAPAILAVGSRFG-LFDPKRAIDVRGW
****::. * *:. .: : .:**:.**:: : : * *::*: *
Mflv_4418|M.gilvum_PYR-GCK RRIGVSVVRWPGPVLAATLALALVGLVTLPGYKTNYDARDYLPEDIPANV
Mvan_1936|M.vanbaalenii_PYR-1 RRIGVSVVRWPGPVLAATLALALVGLVALPGYKTNYDSRDYLPEDVPANV
TH_3260|M.thermoresistible__bu RKVGASVVRWPGPIMVTTVAVTLVGLLALPGYRTNYNDRNYLPASFPSNA
MUL_0757|M.ulcerans_Agy99 RKIGAAIVRWPGPILIGAVALALVGLLTLPGYKTNYNDRNYLPADLPANE
MMAR_0775|M.marinum_M RRVGTAIVRWPGPILAVSIAIALIGLLALPGYRTNYDSKKYLPASTPTNV
Mb0458c|M.bovis_AF2122/97 RRVGTVVVRWPLPVLVATCAIALVGLLALPGYKTSYNDRDYLPDFIPANQ
Rv0450c|M.tuberculosis_H37Rv RRVGTVVVRWPLPVLVATCAIALVGLLALPGYKTSYNDRDYLPDFIPANQ
MLBr_02378|M.leprae_Br4923 RRIGTVVVRWPLPILITTCAIAMVGLLALPGYRTNYKDRAYLPASIPANQ
MAB_4310c|M.abscessus_ATCC_199 RKVGTSVVRWPIPIIVLTSLIAIIGFVSLMTYVPQYNDQKFTPADMPANL
MSMEG_0463|M.smegmatis_MC2_155 RKVGTAVVRWPIPIILVTGLVAAIGFASLLTYVPQYNDQKFTPADMPANV
MAV_4755|M.avium_104 RKVGTAVVRWPKPIIVVTAAIAVIGFISLLTYVPNYDDQKFTPKDMPANA
*::*. :**** *:: : :: :*: :* * ..*. : : * *:*
Mflv_4418|M.gilvum_PYR-GCK GYAVADQHFGSARMNPELLMVESDHDLRNPADFLVIDKIAKSVFRVPGVA
Mvan_1936|M.vanbaalenii_PYR-1 GYAVADRHFGSARMNPELLMVESDHDLRNPADFLVIDKIAKAIFRVPGVA
TH_3260|M.thermoresistible__bu GYEAAERHFDPARLNPELLLIESDHDLRNPADFLVIEKIAKAVFQVEGIS
MUL_0757|M.ulcerans_Agy99 GYSAAERHFSQARMNPEVLMVESDHDMRNSSDFLVINKIAKAIFGVEGIS
MMAR_0775|M.marinum_M GYTAADRHFSQARMNPELLMVETDHDMRNPADMLVIDRIAKGVFHLPGVA
Mb0458c|M.bovis_AF2122/97 GYAAADRHFSQARMKPEILMIESDHDMRNPADFLVLDKLAKGIFRVPGIS
Rv0450c|M.tuberculosis_H37Rv GYAAADRHFSQARMKPEILMIESDHDMRNPADFLVLDKLAKGIFRVPGIS
MLBr_02378|M.leprae_Br4923 GFAAADRHFPQARMKPEILMIESDHDMRNPADFLILDKLARGIFRVPGIS
MAB_4310c|M.abscessus_ATCC_199 AMEVADRHFSQARMNPELLMLEADHDLRTPADMLVIDRVAKGIVHMRGIE
MSMEG_0463|M.smegmatis_MC2_155 AMAVADRHFSEARMNPELLMLEADHDIRNPADMLVIDRVAKYVVHMRGIE
MAV_4755|M.avium_104 AMTAAERHFSQARMNPELLMVEADHDLRDPADMLVVDRIAKSVFHLRGIE
. .*::** **::**:*::*:***:* .:*:*:::::*: :. : *:
Mflv_4418|M.gilvum_PYR-GCK RVQTITRPDGKPIKHTTIPFQMSMQGTTQRLNEKYLQDRMADMLVQADAM
Mvan_1936|M.vanbaalenii_PYR-1 RVQTITRPDGKPIKHTTIPFQMSMRSTTSRLNEKYMQDRMADMLVQADAM
TH_3260|M.thermoresistible__bu RVQGITRPEGTPIEHSSIPFMISQQGTLQILNQQYMQDRMADMLVQADEM
MUL_0757|M.ulcerans_Agy99 RVQAITRPDGKPIEHTSIPFLISMQGTSQKLTEKYNQDLTTSMLQQVNDI
MMAR_0775|M.marinum_M RVQAVTRPLGKPIEHSSIPFQISMQNTVQVENMQYMKQRMADMLIQAAAM
Mb0458c|M.bovis_AF2122/97 RVQAITRPEGTTMDHTSIPFQISMQNAGQLQTIKYQRDRANDMLKQADEM
Rv0450c|M.tuberculosis_H37Rv RVQAITRPEGTTMDHTSIPFQISMQNAGQLQTIKYQRDRANDMLKQADEM
MLBr_02378|M.leprae_Br4923 RVQAITRPDGTAMDHTSIPFQISMQNAGQVQTMKYQKDRMNDLLRQAENM
MAB_4310c|M.abscessus_ATCC_199 RVQTITRPLGAPIEHSSIPFLMGAQNAGTLQTAKFNNENSAQMLEQADEL
MSMEG_0463|M.smegmatis_MC2_155 RVQTITRPLGSPIEHSSIPFLLGAQNAGTMQAAKFNNDNTAQMLEQADEL
MAV_4755|M.avium_104 RVQTITRPLGAPIEHSSIPFQIAMQNSGTLQTAKFMNDTMANMLEQADEL
*** :*** * .:.*::*** :. :.: :: .: .:* *. :
Mflv_4418|M.gilvum_PYR-GCK QTNVDTMTRMQSLMGQMSATTHEMVLKTKKTEIDIIEVRDNLADFDDFFR
Mvan_1936|M.vanbaalenii_PYR-1 QTNVDTMTKMQSLMTQMSAATHDMVTKTKKTAVDIVEVRDYLANLDDFVR
TH_3260|M.thermoresistible__bu QLMIDTMDKMIALMEEMVETTDNMVRDMDEMAAHVAEMRDSISEFDDFFR
MUL_0757|M.ulcerans_Agy99 QTNIDQMERMHDLTQQMADVTHQMVIQMKGMVVDVEELRNHIADFDDFFR
MMAR_0775|M.marinum_M QDSIDTLDRMYDIMGKMVETTHDMDGLTHDMVEITDELRDHIADFDDFWR
Mb0458c|M.bovis_AF2122/97 ATTIAVLTRMHSLMAEMASTTHRMVGDTEEMKEITEELRDHVADFDDFWR
Rv0450c|M.tuberculosis_H37Rv ATTIAVLTRMHSLMAEMASTTHRMVGDTEEMKEITEELRDHVADFDDFWR
MLBr_02378|M.leprae_Br4923 AETIASMRRMHQLMALLTENTHHILNDTVEMQKTTSKLRDEIANFDDFWR
MAB_4310c|M.abscessus_ATCC_199 SRTVASMERMYSLMQELTGTMHSMVGRTHEMEATIQEMRDNLANFDDFFR
MSMEG_0463|M.smegmatis_MC2_155 SRTIASMERMYGLTQELTDTTHSMVGRTHDMVEATKEMRDSLADFDDFFR
MAV_4755|M.avium_104 DKTIAVMEHMYGVMKELTATTHSMVGRTHEMVDTTNELRDRIADFDDFFR
: : :* : : . : ::*: ::::*** *
Mflv_4418|M.gilvum_PYR-GCK PVRNYFYWEPHCYDIPVCWTMRSVFDTLDGINPLTDDISELIPDLERLDE
Mvan_1936|M.vanbaalenii_PYR-1 PVRNYFYWEPHCYNIPVCATMRSVFDTLDGINPLTDDIQELIPDLERLDA
TH_3260|M.thermoresistible__bu PIRNYFYWEPHCYNIPVCWAMRSVFDTLDGINTMTDDIQNMLPHMHRLND
MUL_0757|M.ulcerans_Agy99 PIRSYFYWEKHCFDIPVCWSLRSVFDTLDGVDLMTEDINNLLPLMERLDT
MMAR_0775|M.marinum_M PIRSYFYWEKHCYDVPICWSLRSIFDALDGVDKITEKLAALSRDMDRLDI
Mb0458c|M.bovis_AF2122/97 PIRSYFYWEKHCYGIPICWSFRSIFDALDGIDKLSEQIGVLLGDLREMDR
Rv0450c|M.tuberculosis_H37Rv PIRSYFYWEKHCYGIPICWSFRSIFDALDGIDKLSEQIGVLLGDLREMDR
MLBr_02378|M.leprae_Br4923 PIRSYFYWERHCFNIPICWSFRSIFDALDGVDQIDERLNSIVGDIKNMDL
MAB_4310c|M.abscessus_ATCC_199 PLRNYFYWEKHCFDIPICQSLRSIFDVLDSVDVLADQMHGLIADMDHMDQ
MSMEG_0463|M.smegmatis_MC2_155 PMRNYLYWEPHCYDIPVCQSLRSVFDTLDGIGQLTDEMQGLTIDMDRMDE
MAV_4755|M.avium_104 PIRSYFYWEKHCFDIPVCWSMRSLFDALDGIDELSDQISGLTIDLDHMDR
*:*.*:*** **:.:*:* ::**:**.**.:. : : : : : .::
Mflv_4418|M.gilvum_PYR-GCK LMPQLIALLPSQIETMRSMQAMMLRQYQTQKGQLDQNAAMSEDADAMGEA
Mvan_1936|M.vanbaalenii_PYR-1 LMPQLIALLPSQIESMRNMQSMMLTMYQSQKGLQDQMASQSEDADAMGEA
TH_3260|M.thermoresistible__bu LMPQMLTLLPTTVATMKTMRSMLLTMQATNAGLQDQMAEMQENANAMGEA
MUL_0757|M.ulcerans_Agy99 LMPQMTAMMPEMIQTMKSMKAQMLSMHSTQQGLQDQMAAMQEDSSAMGEA
MMAR_0775|M.marinum_M LMPQMRAQIPFQIASMKTMKTMMLTMHSSMSSLYDQMDEMSKNSTAMGKA
Mb0458c|M.bovis_AF2122/97 LMPQMVAQIPPQIEAMENMRTMILTMHSTMTGIFDQMLEMSDNATAMGKA
Rv0450c|M.tuberculosis_H37Rv LMPQMVAQIPPQIEAMENMRTMILTMHSTMTGIFDQMLEMSDNATAMGKA
MLBr_02378|M.leprae_Br4923 LMPQMLEQFPPMIESMESMRTIMLTMHSTMSGIFDQMNELSDNANTMGKA
MAB_4310c|M.abscessus_ATCC_199 LMPQMLPVLRTTIDSMGRMRDFMIATHSTMAGTQAQQQELAKGATEIGLY
MSMEG_0463|M.smegmatis_MC2_155 LMPQMLPILRSTIDSMKTMRDFMIATHSVMAGTQAQQQELAKNATEIGLY
MAV_4755|M.avium_104 LMPQLLADFPKTIDSMKTMRDFMLSTHSTMAGIQAHQQEAAEGSTLMGQY
****: : : :* *: :: . : ..: :*
Mflv_4418|M.gilvum_PYR-GCK FDDSMNDDSFYLPPEAFDNDDFKRGMENFISPDGRSVRFIISHEGDPATV
Mvan_1936|M.vanbaalenii_PYR-1 FDDSMNDDSFYLPPEAFDNDDFKRGIENFISPDGKSVRFIIAHEGDPATV
TH_3260|M.thermoresistible__bu FDQARNDDSFYLPPEAFDNPDFQRGLEQFLSPDGHAVRFIISHQGDPMTP
MUL_0757|M.ulcerans_Agy99 FDSSRNDDSFYLPPEVFNNPDFKRGLEQFLSPDGHAVRFIISHEGDPMSQ
MMAR_0775|M.marinum_M FDAARNDDSFYIPPEVFDNADFKRGLKMFLSPDGHAVRFIISHEGDPAST
Mb0458c|M.bovis_AF2122/97 FDAAKNDDSFYLPPEVFKNKDFQRAMKSFLSSDGHAARFIILHRGDPQSP
Rv0450c|M.tuberculosis_H37Rv FDAAKNDDSFYLPPEVFKNKDFQRAMKSFLSSDGHAARFIILHRGDPQSP
MLBr_02378|M.leprae_Br4923 FDTAKNDDSFYLPPEVFKNTDFKRAMKSFLSSDGHAARFIILHRGDPASV
MAB_4310c|M.abscessus_ATCC_199 FDQAKNDDFFYLPPDVFTNPDFKRGLKMFVSPDGKAVRFIITHQGDPASV
MSMEG_0463|M.smegmatis_MC2_155 FDQAKNDDFFYLPPDVFDNPDFKRGLEMFVSPDGKAVRMIITHQGDPASV
MAV_4755|M.avium_104 FDEAKNDDSFYLPPEVFKNPDFKRGLKMFVSPDGTAVRFIITHQGDPASV
** : *** **:**:.* * **:*.:: *:*.** :.*:** *.*** :
Mflv_4418|M.gilvum_PYR-GCK EGISEVVPIKTAAKEAIKGTPLEGSTVYLAGTAATYKDMSDGAFYDLLIA
Mvan_1936|M.vanbaalenii_PYR-1 EGISRVLPIKTAAKEAIKGTPLEGSTVYLAGTAATYKDMRDGANYDLLIA
TH_3260|M.thermoresistible__bu EGLAKIEPIKKAAVEAIKGTPLEGSRVYLGGTAAVFKDMSDGTKYDLIIA
MUL_0757|M.ulcerans_Agy99 AGINHIDKIKTAAKEAIKGTPLEGSTIYLGGTVAMFKDLSDGNTFDLMIA
MMAR_0775|M.marinum_M EGISHVKPIMDEAKQAIKGTPLEGAKIYLAGTASVYKDMRDGSRWDLLIA
Mb0458c|M.bovis_AF2122/97 EGIKSIDAIRTAAEESLKGTPLEDAKIYLAGTAAVFHDISEGAQWDLLIA
Rv0450c|M.tuberculosis_H37Rv EGIKSIDAIRTAAEESLKGTPLEDAKIYLAGTAAVFHDISEGAQWDLLIA
MLBr_02378|M.leprae_Br4923 AGIASINAIRTAAEEALKGTPLEDTKIYLAGTAAVFKDIDEGANWDLVIA
MAB_4310c|M.abscessus_ATCC_199 EGIDHVRDLKGVVADAVKGTPLSNAKVSIAGTASMYADMQDGVSIDLMIA
MSMEG_0463|M.smegmatis_MC2_155 DGIAHVRDLRDVVADAVKGTPLANAKVSLAGTASLYSDMQDGVVTDLMIA
MAV_4755|M.avium_104 EGIKHVAGVKDAVADAIKGTPLESSKVYLAGTASMYSDMQEGVIIDLLVA
*: : : . :::***** .: : :.**.: : *: :* **::*
Mflv_4418|M.gilvum_PYR-GCK GIAAVTLIFVIMLVITRSVVAAAVIVGTVLLSLGASFGLSVLLWQHIIGL
Mvan_1936|M.vanbaalenii_PYR-1 GIAAVTLIFVIMLIITRSVVAAAVIVGTVLLSLGASFGLSVLLWQHIIGL
TH_3260|M.thermoresistible__bu AIAAVCLIFIIMLILTRAVVAAAVIVGTVTLSLAASFGVSVLIWQYIVGL
MUL_0757|M.ulcerans_Agy99 GISALCLIFIIMLIITRSVVAAAVIVGTVVLSLGASFGLSVLIWQHVLGI
MMAR_0775|M.marinum_M GVAAVSLILIIMLIITRSLVAAVVIVGTVLLSLGASFGLSVLVWQDIFGV
Mb0458c|M.bovis_AF2122/97 AISSLCLIFIIMLIITRAFIAAAVIVGTVALSLGASFGLSVLLWQHILAI
Rv0450c|M.tuberculosis_H37Rv AISSLCLIFIIMLIITRAFIAAAVIVGTVALSLGASFGLSVLLWQHILAI
MLBr_02378|M.leprae_Br4923 GISSLCLIFIIMLIITRAFVAAAVIVGTVALSLGASFGLSVLLWQHILGI
MAB_4310c|M.abscessus_ATCC_199 VIASMILIFAIMVLITRSVVAALVIVGTVAASLGTACGLSVLLWQDILGL
MSMEG_0463|M.smegmatis_MC2_155 IIASMILIFGIMLVITRSVVAALVIVGTVAASLGTACGLSVLLWQDILGL
MAV_4755|M.avium_104 GISCLILIFTIMLIITRSVVAALVIVGTVAASLGTACGLSVLMWQDLIGL
::.: **: **:::**:.:** ****** **.:: *:***:** :..:
Mflv_4418|M.gilvum_PYR-GCK ALHWMVLPMAVILLLAVGSDYNLLLVSRFKEELSGGLKTGIIRAMAGTGS
Mvan_1936|M.vanbaalenii_PYR-1 ALHWMVLPMAVILLLAVGSDYNLLLVSRFKEELAGGLKTGIIRSMAGTGS
TH_3260|M.thermoresistible__bu EMHWMVFPMALIILLAVGADYNLLVVARLREEIPAGLNTGMIRAMGGSGS
MUL_0757|M.ulcerans_Agy99 ELHWLVLAMAVIILLAVGADYNLLLVARLKEEIHAGINTGIIRAMGGSGS
MMAR_0775|M.marinum_M ELHWMVLAMSVILLLAVGSDYNLLLVSRLKEEIGAGLKTGIIRAMAGTGG
Mb0458c|M.bovis_AF2122/97 HLHWLVLAMSVIVLLAVGSDYNLLLVSRFKQEIGAGLKTGIIRSMGGTGK
Rv0450c|M.tuberculosis_H37Rv HLHWLVLAMSVIVLLAVGSDYNLLLVSRFKQEIGAGLKTGIIRSMGGTGK
MLBr_02378|M.leprae_Br4923 ELHYLVLAMSVIVLLAVGSDYNLLLVSRFKQEIQAGLKTGIIRSMGGTGK
MAB_4310c|M.abscessus_ATCC_199 GVQWIVIPLSMVILLAVGSDYNLLVVSRLKEEIHAGLNTGMIRGMGATGR
MSMEG_0463|M.smegmatis_MC2_155 GVQWVVLPLSIVILLAVGSDYNLLLVSRLKEEIHAGLNTGIIRGMGATGR
MAV_4755|M.avium_104 GVQWIVLPLSIVILLAVGSDYNLLLVSRLKEEIPAGLNTGIIRGMGASGR
::::*:.:::::*****:*****:*:*:::*: .*::**:**.*..:*
Mflv_4418|M.gilvum_PYR-GCK VVTSAGLVFAFTMASFAFSDLKVMAQVGTTIALGLLFDTLIVRSFMTPAV
Mvan_1936|M.vanbaalenii_PYR-1 VVTSAGLVFAFTMASFAFSDLKVMAQVGTTIALGLLFDTLIVRSFMTPAV
TH_3260|M.thermoresistible__bu VVTAAGLVFGLTMMVMVVSDLTVMGQVGTTIGLGLLFDTFVVRSFMMPSI
MUL_0757|M.ulcerans_Agy99 VVTAAGLVFAFTMMSFAVSELTVMAQVGTTIGMGLLFDTLIVRSFMTPSI
MMAR_0775|M.marinum_M VVTTAGLVFAATMASFIFSDLRVIGQVGTTIGLGLLFDTLIVRSFMTPSI
Mb0458c|M.bovis_AF2122/97 VVTNAGLVFAVTMASMAVSDLRVIGQVGTTIGLGLLFDTLIVRSFMTPSI
Rv0450c|M.tuberculosis_H37Rv VVTNAGLVFAVTMASMAVSDLRVIGQVGTTIGLGLLFDTLIVRSFMTPSI
MLBr_02378|M.leprae_Br4923 VVTNAGLVFAFTMASMVVSDLRVIGQVGTTIGLGLLFDTLIVRSFMMPSI
MAB_4310c|M.abscessus_ATCC_199 VVTAAGLVFAFTMMSMIVSDLRVVGQLGMTIGIGLIVDTLIVRAFMTPSI
MSMEG_0463|M.smegmatis_MC2_155 VVTAAGLVFAFTMMAMIVSDLRVVGQLGTTIGLGLIVDTLIVRAFMTPSI
MAV_4755|M.avium_104 VVTAAGLVFAFTMASMIVSQLRVIGELGTTIALGLLVDTLIVRSFMTPSI
*** *****. ** : .*:* *:.::* **.:**:.**::**:** *::
Mflv_4418|M.gilvum_PYR-GCK AALLGRWFWWPQKFRSAASERRLARIGVQPATAGR---------------
Mvan_1936|M.vanbaalenii_PYR-1 AALLGRWFWWPQKVRSHASQRRLAALNH----------------------
TH_3260|M.thermoresistible__bu ATLLGRWFWWPQFVRKRPVPQPWPSVPPRGPATTDRVSV-----------
MUL_0757|M.ulcerans_Agy99 AALLGKWFWWPQVVRQRPAPQPWPTPAPTPPPATTSV-------------
MMAR_0775|M.marinum_M AALMGRWFWWPQQVRTRPASQLLRPYGPRPLVRALLLPRNGDSADGPEDT
Mb0458c|M.bovis_AF2122/97 AALLGRWFWWPLRVRSRPARTPTVPSETQPAGRPLAMSSDRLG-------
Rv0450c|M.tuberculosis_H37Rv AALLGRWFWWPLRVRSRPARTPTVPSETQPAGRPLAMSSDRLG-------
MLBr_02378|M.leprae_Br4923 AALLGRWFWWPQQGRTRPLLTVAAPVGLPPD-RSLVYSD-----------
MAB_4310c|M.abscessus_ATCC_199 AAALGRWFWWPINVFEIVRRGRETQAPEPVTAPLPVVDAGPQPPTAPSTS
MSMEG_0463|M.smegmatis_MC2_155 AAALGRWFWWPLNTFEITRRGR--QAPHARS-----VDGNTQPFPGVTTA
MAV_4755|M.avium_104 AAALGHWFWWPINTIRMTRRTHHDEEAHTAP--------IRQPALR----
*: :*:*****
Mflv_4418|M.gilvum_PYR-GCK ------------
Mvan_1936|M.vanbaalenii_PYR-1 ------------
TH_3260|M.thermoresistible__bu ------------
MUL_0757|M.ulcerans_Agy99 ------------
MMAR_0775|M.marinum_M NTERLAVSSPHY
Mb0458c|M.bovis_AF2122/97 ------------
Rv0450c|M.tuberculosis_H37Rv ------------
MLBr_02378|M.leprae_Br4923 ------------
MAB_4310c|M.abscessus_ATCC_199 ------------
MSMEG_0463|M.smegmatis_MC2_155 P-----------
MAV_4755|M.avium_104 ------------