For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLLTITKLTDSVGAEVTGLDPAALAHDDSVGEAVLDALEDNGVLVFRGLYLDPAAQVAFCGRLGEVDHSS DGHHPVPGIYPITLDKSKNASAAYLKATFDWHIDGCTPLGDECPQKATVLSAVRVAERGGETEFANSYGA YDALTDDEKQRFGALRVVHSLEASQRRVYPDPSPELVARWRSRRTHEHPLVWTHRSGRKSLVLGASADYV VGMDVDEGRALLEELLQRATVPERVYRHGWSIGDTVIWDNRGVLHRAAPYDPDSSREMLRTTVLGDEPIQ *
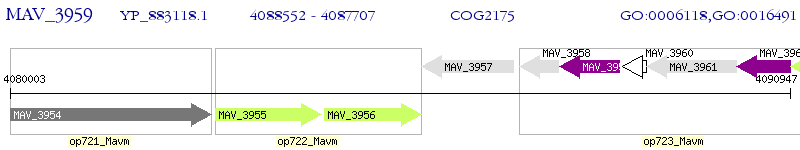
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3959 | - | - | 100% (281) | taurine catabolism dioxygenase TauD, TfdA family protein |
| M. avium 104 | MAV_0235 | - | 7e-26 | 32.36% (275) | taurine catabolism dioxygenase TauD/TfdA |
| M. avium 104 | MAV_4353 | - | 2e-23 | 31.21% (282) | alpha-ketoglutarate-dependent taurine dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3440 | - | 1e-25 | 33.00% (300) | dioxygenase |
| M. gilvum PYR-GCK | Mflv_2472 | - | 1e-134 | 81.85% (281) | taurine catabolism dioxygenase TauD/TfdA |
| M. tuberculosis H37Rv | Rv3406 | - | 1e-25 | 33.00% (300) | dioxygenase |
| M. leprae Br4923 | MLBr_01992 | - | 7e-05 | 22.58% (279) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0086 | - | 1e-21 | 28.08% (292) | putative taurine dioxygenase |
| M. marinum M | MMAR_1141 | tauD | 8e-22 | 31.38% (290) | taurine catabolism dioxygenase, TauD |
| M. smegmatis MC2 155 | MSMEG_0291 | - | 1e-126 | 76.51% (281) | dioxygenase, TauD/TfdA family protein |
| M. thermoresistible (build 8) | TH_0024 | - | 1e-129 | 78.65% (281) | dioxygenase, TauD/TfdA family protein |
| M. ulcerans Agy99 | MUL_0904 | tauD | 5e-22 | 31.38% (290) | taurine catabolism dioxygenase, TauD |
| M. vanbaalenii PYR-1 | Mvan_4182 | - | 1e-130 | 79.36% (281) | taurine catabolism dioxygenase TauD/TfdA |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3440|M.bovis_AF2122/97 MTDLITVKKLGSRIGAQIDGVRLGGDLDPAAVNEIRAALLAHKVVFFRGQ
Rv3406|M.tuberculosis_H37Rv MTDLITVKKLGSRIGAQIDGVRLGGDLDPAAVNEIRAALLAHKVVFFRGQ
MMAR_1141|M.marinum_M MTDLITVKKLGSRIGAQVDGVSLGADLDAAAVDQIRAALLEHKVIFFRNQ
MUL_0904|M.ulcerans_Agy99 MTDLITVKKLGSRIGAQVDGVSLGADLDAAAVDQIRAALLEHKVIFFRNQ
MAB_0086|M.abscessus_ATCC_1997 MSSDLRVVKLGENIGARIDGVRLG-EVDTATASAINEAMLEHKVVFFRGQ
MAV_3959|M.avium_104 -MSLLTITKLTDSVGAEVTGLDPAALAHDDSVGEAVLDALEDNGVLVFRG
Mflv_2472|M.gilvum_PYR-GCK -MSLLTINKLTPSVGAEVLGVDPVRLASDESLAAAVLDALEDNGVLVFPD
TH_0024|M.thermoresistible__bu -MSRLTINRLTASVGAEVTGTTADELAADAALGAAVLDALEEHGVLVFRG
Mvan_4182|M.vanbaalenii_PYR-1 -MTLLTINKLTSSVGAEVTGLDPGRLAGDDALGTAVLDALEDNGVLVFPG
MSMEG_0291|M.smegmatis_MC2_155 -MSVLTINKLTASVGAEVVGIDSERLATDDGIAAAVLDALEDNGVLVFRG
MLBr_01992|M.leprae_Br4923 ----MTLDVKGDGLGAQVTGVDPKNLDSISTAEIRKIVYVN--KLVVLKD
: : :**.: * : :..
Mb3440|M.bovis_AF2122/97 HQLDDAEQLAFAGLLGTPIGHPAAIALADDAPIITPINSEFGK-----AN
Rv3406|M.tuberculosis_H37Rv HQLDDAEQLAFAGLLGTPIGHPAAIALADDAPIITPINSEFGK-----AN
MMAR_1141|M.marinum_M HHLDDQQQLQFAGLLGTPIGHPAAAAALPDAPIITPINSEWGK-----AN
MUL_0904|M.ulcerans_Agy99 HHLDDQQQLQFAGLLGTPIGHPAAAAALPDAPIITPINSEWGK-----AN
MAB_0086|M.abscessus_ATCC_1997 HHVDDEVQFAFAKSMGIPT-TPHPTLTSSDVKVLPIDSEEGGR-----AN
MAV_3959|M.avium_104 LYLDPAAQVAFCGRLGEVDHSSDGHHPVPGIYPITLDKSKNAS-----AA
Mflv_2472|M.gilvum_PYR-GCK LHLAPEAQVAFSRRLGAVDHSADGHHPVAGIYPVTLDKSKNAS-----AA
TH_0024|M.thermoresistible__bu LRIDPQTQVKFCRHLGEVDHSSDGHHPVAGIYPVTLDKSKNSS-----AA
Mvan_4182|M.vanbaalenii_PYR-1 LHLEPEVQVEFCRRLGEIDFSSDGHHRVAGIYPVTLDKSKNSS-----AA
MSMEG_0291|M.smegmatis_MC2_155 LYLEPEAQVAFSQRLGEVDRSADGHHPVSGIYPITLDQTKNKA-----AE
MLBr_01992|M.leprae_Br4923 VHPTPEEFIKLGRIIGEIVPYYEPIYRHKDYPEIFVSSTEEGQGVPNTGA
. : :* . : . : .
Mb3440|M.bovis_AF2122/97 RWHTDVTFAAN---------YPAASVLRAVSLPSYGGSTLWANTAAAYAE
Rv3406|M.tuberculosis_H37Rv RWHTDVTFAAN---------YPAASVLRAVSLPSYGGSTLWANTAAAYAE
MMAR_1141|M.marinum_M RWHTDVTFAAN---------YPAASILRAVTLPNYGGSTLWANTATAYAE
MUL_0904|M.ulcerans_Agy99 RWHTDVTFAAN---------YPAASILRAVTLPNYGGSTLWANTATAYAE
MAB_0086|M.abscessus_ATCC_1997 QWHTDVTFVDR---------IPKASILRAVELPPYGGTTTWTSTVAAYRQ
MAV_3959|M.avium_104 YLKATFDWHIDGCTPLGDECPQKATVLSAVRVAERGGETEFANSYGAYDA
Mflv_2472|M.gilvum_PYR-GCK YLRATFDWHIDGCTPIGDECPQRATVLSAVQVADRGGETEFANSYAAYEA
TH_0024|M.thermoresistible__bu YLRATFDWHIDGCTPTGDEYPQKATVLSAVQVADRGGETEFASSYGAYEA
Mvan_4182|M.vanbaalenii_PYR-1 YLRATFDWHIDGCTPTGDDYPQMATVLSAKQVAESGGETEFASSYAAYDA
MSMEG_0291|M.smegmatis_MC2_155 YLKATFDWHIDGCTPLNDECPQKATVLSAIEVAARGGETEFANAYAAYES
MLBr_01992|M.leprae_Br4923 FWHVDYIFMPE---------PFAFSMTLPLAMPGHDRGTHFIDLSQVWES
:. : :: . :. . * : . .:
Mb3440|M.bovis_AF2122/97 LPEPLKCLTENLWALHTNRYDYVT---TKPLTAAQRA-----FRQVFEKP
Rv3406|M.tuberculosis_H37Rv LPEPLKCLTENLWALHTNRYDYVT---TKPLTAAQRA-----FRQVFEKP
MMAR_1141|M.marinum_M LPEPLKCLVENLWALHTNRYDYVANEAVQALTDTQQA-----FRQAFQKP
MUL_0904|M.ulcerans_Agy99 LPEPLKCLVENLWALHTNRYDYVANEAVQALTDTQQA-----FRQAFQKP
MAB_0086|M.abscessus_ATCC_1997 LPKPLQDLADNLWAMHNNQFDYTQVDPAKVAELLAKAGSGSKYVREFGAT
MAV_3959|M.avium_104 LTDDEKQRFGALRVVHSLEASQRRVYPDPSPELVARWR-----------S
Mflv_2472|M.gilvum_PYR-GCK FSDEEKERYAALRVVHSLEASQSRVNPDPSPEELARWR-----------S
TH_0024|M.thermoresistible__bu LSEEEKRHVETLRVVHSLEASQRRVTPDPTPEQLARWR-----------A
Mvan_4182|M.vanbaalenii_PYR-1 LTDAEKQRFASLRVVHSLEASQRRVNPDPSPEELARWR-----------A
MSMEG_0291|M.smegmatis_MC2_155 LDADEKQRYARLRVVHSLEASQRRVYPDPTAEQRRRWA-----------A
MLBr_01992|M.leprae_Br4923 LPNAMKDSVRGTYSNHSPRR-YIKIRPSDVYRPVGEVL------AEIEEV
: : *. . .
Mb3440|M.bovis_AF2122/97 DFRTEHPVVRVHPETGERTLLAGDFVRSFVGLDSH--ESRVLFEVLQRRI
Rv3406|M.tuberculosis_H37Rv DFRTEHPVVRVHPETGERTLLAGDFVRSFVGLDSH--ESRVLFEVLQRRI
MMAR_1141|M.marinum_M DFRTEHPVVRVHPETGERTLLAGDFVRGFVGLDSQ--ESSALFELLQRRI
MUL_0904|M.ulcerans_Agy99 DFRTEHPVVRVHPETGERTLLAGDFVRGFVGLDSQ--ESSALFELLQRRI
MAB_0086|M.abscessus_ATCC_1997 HFETHHPVVRVHPETGEKALLLGNFVKRILDVSAS--ESQALFRMFQDRV
MAV_3959|M.avium_104 RRTHEHPLVWTH-RSGRKSLVLGASADYVVGMDVD--EGRALLEELLQRA
Mflv_2472|M.gilvum_PYR-GCK RPTHEHPLVWTH-RSGRKSLVLGASAHYVVGMDYD--EGRALLAELLDRA
TH_0024|M.thermoresistible__bu RPTHEHPLVWTH-RNGRKSLVLGASADYVVGMDLD--EGRALLADLLDRA
Mvan_4182|M.vanbaalenii_PYR-1 RPTHEHPLVWTH-RTGRKSLVLGASADYIVGMDPE--EGRALLSELLDRA
MSMEG_0291|M.smegmatis_MC2_155 RPTHENPLVWTH-RNGRKSLVLGASADHIVGMDRD--EGRALLDGLLART
MLBr_01992|M.leprae_Br4923 TPPQKWPTVIKHPKTGQEILYICEAATVSVEDKNGNLLDPMVLQELLTAS
. * * * ..*.. * . : . . :: :
Mb3440|M.bovis_AF2122/97 TMPE------NTIRWNWAPGDVAIWDNRATQHRAIDDYDDQHRLMHRVTL
Rv3406|M.tuberculosis_H37Rv TMPE------NTIRWNWAPGDVAIWDNRATQHRAIDDYDDQHRLMHRVTL
MMAR_1141|M.marinum_M TSPE------NTIRWNWESGDVAIWDNRATQHRAIDDYDDQHRLLHRVTL
MUL_0904|M.ulcerans_Agy99 TSPE------NTIRWNWESGDVAIWDNRATQHRAIDDYDDQHRLLHRVTL
MAB_0086|M.abscessus_ATCC_1997 TWLE------NTIRWSWELGDVAMWDNRATQHYAISDYGDQPRRMHRVTL
MAV_3959|M.avium_104 TVPE------RVYRHGWSIGDTVIWDNRGVLHRAAPYDPDSSREMLRTTV
Mflv_2472|M.gilvum_PYR-GCK TQPE------LVYSHSWSVGDTVIWDNNGVLHRAAPYDPDSQREMLRTTV
TH_0024|M.thermoresistible__bu TTED------RVYRHRWSVGDTVIWDNRGVLHRAAPYPEDSPREMLRTTV
Mvan_4182|M.vanbaalenii_PYR-1 TTAD------KVYSHHWAVGDTVIWDNRGVLHRAARYDENSPREMLRTTV
MSMEG_0291|M.smegmatis_MC2_155 TTPD------KVYSHKWSVGDTVIWDNQGVLHRAAPYEPDSPRHMLRTTV
MLBr_01992|M.leprae_Br4923 GQLDPDCKSLLIHTQHYEVGDVVLWDNRALVHRAKHGTVSGTLITYRLTL
: : **..:***.. * * . * *:
Mb3440|M.bovis_AF2122/97 MGDVPVDVYGQASRVISG---------APMEIAG
Rv3406|M.tuberculosis_H37Rv MGDVPVDVYGQASRVISG---------APMEIAG
MMAR_1141|M.marinum_M MGDVPVDVHGQRSRVISG---------APLALAG
MUL_0904|M.ulcerans_Agy99 MGDVPVDVHGQRSRVISG---------APLALAG
MAB_0086|M.abscessus_ATCC_1997 AGDVPVGVNGESSRVIAGDASDYSVIDNPKRLVA
MAV_3959|M.avium_104 LGDEPIQ---------------------------
Mflv_2472|M.gilvum_PYR-GCK LGDEPIQ---------------------------
TH_0024|M.thermoresistible__bu LGDEPIQ---------------------------
Mvan_4182|M.vanbaalenii_PYR-1 LGDEPIQ---------------------------
MSMEG_0291|M.smegmatis_MC2_155 LGDEPIR---------------------------
MLBr_01992|M.leprae_Br4923 LDGLKTPGYAV-----------------------
..