For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGQRCDGMVALVTGSSRGLGRAIAARLAERGATVALTARTLDPDPKYQGALRQTRDEILAAGGKAVAVQA DLSQPDERERLFAEVVGTVGAPDILVNNAAVTFLRPLDGFPQRRARLMMEMHVLGPLHLCQLAIPAMRER GRGWIVNLTSVGGDLPPGPPFSEFDRTAGFGIYGTAKAALNRLTKSLAAELYDDGIAVNAAAPSNPVATP GAGTLDLAKTDTEDIALITETVFRLCTGDPKTLTGRIAHTQPFLAEVGWPGTGPPAT*
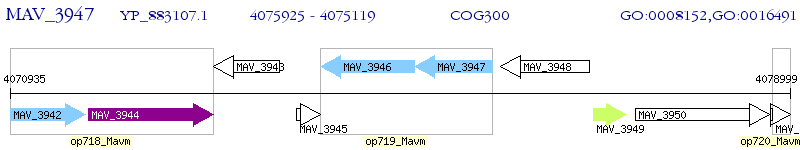
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3947 | - | - | 100% (268) | NAD dependent epimerase/dehydratase family protein |
| M. avium 104 | MAV_3027 | - | 1e-42 | 40.80% (250) | short-chain dehydrogenase/reductase SDR |
| M. avium 104 | MAV_1815 | - | 3e-28 | 31.76% (255) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1079 | - | 5e-24 | 35.44% (206) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4556 | - | 2e-44 | 43.60% (250) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1050 | - | 5e-24 | 35.44% (206) | oxidoreductase |
| M. leprae Br4923 | MLBr_01740 | - | 7e-16 | 29.33% (225) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3946c | - | 3e-22 | 29.57% (257) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_1395 | - | 2e-31 | 37.97% (266) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2885 | - | 5e-47 | 42.80% (250) | short-chain dehydrogenase/reductase SDR |
| M. thermoresistible (build 8) | TH_0319 | - | 1e-34 | 42.00% (200) | short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4828 | - | 4e-23 | 37.24% (196) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_2884 | - | 4e-45 | 42.00% (250) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2885|M.smegmatis_MC2_155 -----------------------------MTN-LLSGRTALVTGSSRGIG
TH_0319|M.thermoresistible__bu --------------------------------------------------
Mvan_2884|M.vanbaalenii_PYR-1 -----------------------------MSG-ILSGKTALVTGSSRGIG
Mflv_4556|M.gilvum_PYR-GCK -----------------------------MTG-LLTGRTALVTGSSRGIG
MAV_3947|M.avium_104 -----------------------------MSGQRCDGMVALVTGSSRGLG
MMAR_1395|M.marinum_M -----------------------------MTG-ALAGRAAIVTGASRGLG
Mb1079|M.bovis_AF2122/97 -----------------------------MARQRFRDQVVLITGASSGIG
Rv1050|M.tuberculosis_H37Rv -----------------------------MARQRFRDQVVLITGASSGIG
MAB_3946c|M.abscessus_ATCC_199 -----------------------------MGS--LAGKTIIMSGGSRGIG
MUL_4828|M.ulcerans_Agy99 MNLATQALAKAFERASNGLANPATLADPDRLRGAVTGKTALVTGASYGIG
MLBr_01740|M.leprae_Br4923 ---------------------------MVKTAQYFVGKRCFVTGAASGIG
MSMEG_2885|M.smegmatis_MC2_155 RAVAQRLAAEGAVVAVTARSYEPSPSVRAGQATALPGTIGETIELIEAAG
TH_0319|M.thermoresistible__bu ---------------------------------VLPGTIGETIELIEAAG
Mvan_2884|M.vanbaalenii_PYR-1 RAVAQRLAAEGATVAVTARSYQPSVSTRAGKDAELPGTIGETVELIEASG
Mflv_4556|M.gilvum_PYR-GCK RAIAQRLAAEGATVAVTARAHQPSESVRAGVSAALPGTIDETIALIEQSG
MAV_3947|M.avium_104 RAIAARLAERGATVALTARTLDP--------DPKYQGALRQTRDEILAAG
MMAR_1395|M.marinum_M RAIALALAAEGAAVVVAGRTEQVW-------DSRLPGTIGDTVAQIEAAG
Mb1079|M.bovis_AF2122/97 EATAKAFAREGAVVALAAR---------------REGALRRVAREIEAAG
Rv1050|M.tuberculosis_H37Rv EATAKAFAREGAVVALAAR---------------REGALRRVAREIEAAG
MAB_3946c|M.abscessus_ATCC_199 LAIALRAARDGANVAIMAKTAEP--------HPKLPGTIYTAAEEIEAAG
MUL_4828|M.ulcerans_Agy99 EATARKLAAAGATVLMVARS---------------AERLEDLVSAIAAGG
MLBr_01740|M.leprae_Br4923 RATALRLAAYGAELYLTDR--------------HCDGLAQTVSEARALGA
..
MSMEG_2885|M.smegmatis_MC2_155 GKAFGIAADLEDAEQRAALVDQVIERTGRIDILVNNAGFAD---------
TH_0319|M.thermoresistible__bu GKAFGIVADLEDPRQRADLIDQVVQHTGRIDILVNNAGFAD---------
Mvan_2884|M.vanbaalenii_PYR-1 GMAFGIACDLEIAGQRASLVDQVVERTGRLDILVNNAGYAD---------
Mflv_4556|M.gilvum_PYR-GCK GTAFGLAADLENSEDRAALVDRVVERTGRVDILVNNAGFAD---------
MAV_3947|M.avium_104 GKAVAVQADLSQPDERERLFAEVVGTVGAPDILVNNAAVTF---------
MMAR_1395|M.marinum_M GRAVAVRADLTDRDDVATLVTGARDSLGPITILVNNAAFTAPGRPPVPGA
Mb1079|M.bovis_AF2122/97 GRAMVAPLDVSSSESVRAMVADVVGEFGRIDVVFNNAGVSLVG-------
Rv1050|M.tuberculosis_H37Rv GRAMVAPLDVSSSESVRAMVADVVGEFGRIDVVFNNAGVSLVG-------
MAB_3946c|M.abscessus_ATCC_199 GHALPLLGDVRDDEVVASAVAQTVEKFGGIDIVVNNASALN---------
MUL_4828|M.ulcerans_Agy99 GAAVAYPTDLTDEAAVGVLTKQVNQNHGPLDIVVSNAGKSLRR-------
MLBr_01740|M.leprae_Br4923 QVPEYRALDISDYDEVAAFGADIHARHPSMDIVMNIAGVSVWG-------
. *: ::.. *.
MSMEG_2885|M.smegmatis_MC2_155 ---------------YSIIEDMPMDTFDRTVEHYLRTPFVLTQAAVPHMR
TH_0319|M.thermoresistible__bu ---------------YSMVEKMSLETFDRTVEHYLRTPFVLTQAAIPHMR
Mvan_2884|M.vanbaalenii_PYR-1 ---------------YSIIEDMPMETFDRTVEHYVRTPFVLTQKAVPHMR
Mflv_4556|M.gilvum_PYR-GCK ---------------YSVVADMSMATFERTVEHYVRTPFVLTQAAVPHMK
MAV_3947|M.avium_104 ---------------LRPLDGFPQRRARLMMEMHVLGPLHLCQLAIPAMR
MMAR_1395|M.marinum_M APRAKSSRTAVGKPGWPGFVSVPLAAYRRHFETSVFAAYELMQLSCPDMI
Mb1079|M.bovis_AF2122/97 ----------------PVDAETFLDDTREMLEIDYLGTVRVVREVLPIMK
Rv1050|M.tuberculosis_H37Rv ----------------PVDAETFLDDTREMLEIDYLGTVRVVREVLPIMK
MAB_3946c|M.abscessus_ATCC_199 ---------------LAPSESISMKAYDLMQDINARGSFSLSTSAIGALK
MUL_4828|M.ulcerans_Agy99 ---------------SLHDQYDRPHDFQRTIDINYLGPIWLLLGLLPAMR
MLBr_01740|M.leprae_Br4923 -----------------TVDRLTHDQWSNVVAINLMGPIHIIETFVPAIL
. : :
MSMEG_2885|M.smegmatis_MC2_155 NQGAG-WIVNIGSVTG----------------LAPVRPYREYNKSSGDVI
TH_0319|M.thermoresistible__bu AQGAG-WIVNIGSVTG----------------LPPIRPYRDYNKTSGDVI
Mvan_2884|M.vanbaalenii_PYR-1 RQGAG-WIVNIGSVTG----------------LAPARPYRDYNKTSGDVV
Mflv_4556|M.gilvum_PYR-GCK TQGQG-WIVNIGSVTG----------------LAPARPYREYNKTSGDVI
MAV_3947|M.avium_104 ERGRG-WIVNLTSVGGD---------------LPPGPPFSEFDRTAGFGI
MMAR_1395|M.marinum_M AAGAG-SIINITSVASR---------------LPGDGPYPDRS-GGVLPG
Mb1079|M.bovis_AF2122/97 QQRSG-RIMNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQELRGSGIA
Rv1050|M.tuberculosis_H37Rv QQRSG-RIMNMSSVVGRKAFARFAGYSSAMHAIAGFSDALRQELRGSGIA
MAB_3946c|M.abscessus_ATCC_199 EADNP-HILTLS----------------------PPITLEPKWFERTTTA
MUL_4828|M.ulcerans_Agy99 ESGRG-HVVNVSSVG------------------------VRLVPGPQWGA
MLBr_01740|M.leprae_Br4923 AAGRGGHLVNVSSAAGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIG
::.:
MSMEG_2885|M.smegmatis_MC2_155 YASMKAALHRFTQGVAAELLDANIAVNAVG------PSSAVRTPGAASLI
TH_0319|M.thermoresistible__bu YASLKAALHRFTQGVAAELLDSNIAVNAVG------PSSAVRTPGAASLI
Mvan_2884|M.vanbaalenii_PYR-1 YASMKAALHRFTQGVAAELLDSNIAVNVVG------PSSAVRTPGAASLI
Mflv_4556|M.gilvum_PYR-GCK YASMKAALHRFTQGVAAELLDADIAVNAVG------PSSAVRTPGAAALI
MAV_3947|M.avium_104 YGTAKAALNRLTKSLAAELYDDGIAVNAAA------PSNPVATPGAGTLD
MMAR_1395|M.marinum_M YGGSKAALEHLTQCAAFDLADHHIAVNALA------PSKPIPTPGLSYYA
Mb1079|M.bovis_AF2122/97 VSVIHPALTQTPLLANVDPADMPPPFRSLT------PIPVHWVAAAVLDG
Rv1050|M.tuberculosis_H37Rv VSVIHPALTQTPLLANVDPADMPPPFRSLT------PIPVHWVAAAVLDG
MAB_3946c|M.abscessus_ATCC_199 YTISKFSMSLVAIGLAAELRQYGIASNALW------PRTTIDTAAIRNIL
MUL_4828|M.ulcerans_Agy99 YQASKGAFDRWLRSVAPELHGDGVDVTSVY------FALVRTRMIEPTPM
MLBr_01740|M.leprae_Br4923 VSVVVPGAVRTPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERAAEKI
. :
MSMEG_2885|M.smegmatis_MC2_155 PDTFPT--EPVEYLAETVLAMCHRPAAERTGLVAFSLHYPWSQGLAVHSL
TH_0319|M.thermoresistible__bu PDTFPT--EPVEYLAETVLAMCHRPAAERTGLVAFSLHYPWSQRIPVHTL
Mvan_2884|M.vanbaalenii_PYR-1 PDTFPT--EPVEYLAETVLAMCHLPAVERTGLVAFSLHYPWSHRLTVHSL
Mflv_4556|M.gilvum_PYR-GCK PDSFPT--EPVEYLAETVLAMCHRPAAERTGLVAFSLHYPWSQKLPVHSL
MAV_3947|M.avium_104 LAKTDT--EDIALITETVFRLCTGDPKTLTGRIAHTQPF-----LAEVGW
MMAR_1395|M.marinum_M REFDDT--ASAEEFARAAVVLASVDADLVTGRTIG-HHEVLDGSFRAFGR
Mb1079|M.bovis_AF2122/97 VARRRA--RVVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYR
Rv1050|M.tuberculosis_H37Rv VARRRA--RVVVPFQPRLLMVGDAFSPRYGDRVVRLLESKIFGRLIGSYR
MAB_3946c|M.abscessus_ATCC_199 GAELVARCRSAQIMADAAYEILTKPSREVSGNCFIDDEVLREAGVTDFSQ
MUL_4828|M.ulcerans_Agy99 LGRLPG-LEPDQAADAVAKAIVERPRTNEPPWLWPAELASVLLAGPAEQA
MLBr_01740|M.leprae_Br4923 LAGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRALRPSP
MSMEG_2885|M.smegmatis_MC2_155 DGRDLLPPLEPPASANPNILPAGI---
TH_0319|M.thermoresistible__bu DGREVLPPLEPPATANPNILPGGM---
Mvan_2884|M.vanbaalenii_PYR-1 DGTTTLPPLEPPANANPNILPLGL---
Mflv_4556|M.gilvum_PYR-GCK DGATVLPPLEPPATANPNILPAGL---
MAV_3947|M.avium_104 PGT------GPPAT-------------
MMAR_1395|M.marinum_M D--------------------------
Mb1079|M.bovis_AF2122/97 GSVYRHQPTESAKAQAAQPERGYSSAR
Rv1050|M.tuberculosis_H37Rv GSVYRHQPTESAKAQAAQPERGYSSAR
MAB_3946c|M.abscessus_ATCC_199 YRECAEEDLELDFWMERA---------
MUL_4828|M.ulcerans_Agy99 ARVWHRRFFAPTAEAGKS---------
MLBr_01740|M.leprae_Br4923 ATMRCVEPIKVGVYSQPRSGEGGKSGN