For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GALSPRTLKDRVVVVTGASRGIGREIAIRAAADGAAVALLAKTQTPNPKIAGTLTETAEAVRTAGGRALP LAVDVRDADGVASAVAEAADEFGGIDVVVNNAGALDLRPTPKLPPKNFHRLLGVNVEGPFAVVQAAYTYL RNSDNAHIVNISPPLNLDPRWVGAHVGHTVGKYAESLLTLGWAAEFASIPVAVNSLWPATTVASTGMIVA MGEDVVRAQARNTQIMADAVHALVTRPASDCSGHFYTDEEILREEGWDDDDLSEYLLAAREEDLTPNFYL SSTPLPSV*
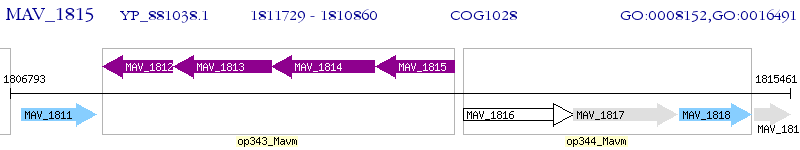
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1815 | - | - | 100% (289) | short-chain dehydrogenase/reductase SDR |
| M. avium 104 | MAV_2555 | - | e-125 | 80.14% (277) | short-chain dehydrogenase/reductase SDR |
| M. avium 104 | MAV_0855 | - | 2e-61 | 44.75% (257) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3251 | - | 2e-53 | 40.71% (253) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2386 | - | 3e-56 | 40.98% (266) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv3224 | - | 2e-53 | 40.71% (253) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 9e-14 | 28.97% (214) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3946c | - | 7e-61 | 42.91% (275) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_1333 | - | 2e-51 | 39.53% (253) | iron-regulated short-chain dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_1894 | - | 1e-55 | 42.06% (252) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_2687 | - | 1e-110 | 77.82% (257) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_2546 | - | 4e-51 | 39.13% (253) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4262 | - | 7e-55 | 42.23% (251) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1815|M.avium_104 --------------------------------------------------
TH_2687|M.thermoresistible__bu --------------------------------------------------
Mb3251|M.bovis_AF2122/97 --------------------------------------------------
Rv3224|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1333|M.marinum_M --------------------------------------------------
MUL_2546|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1894|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3946c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2386|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4262|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
MAV_1815|M.avium_104 --------------------------------------------------
TH_2687|M.thermoresistible__bu --------------------------------------------------
Mb3251|M.bovis_AF2122/97 --------------------------------------------------
Rv3224|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1333|M.marinum_M --------------------------------------------------
MUL_2546|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1894|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3946c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2386|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4262|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
MAV_1815|M.avium_104 --------------------------------------------------
TH_2687|M.thermoresistible__bu --------------------------------------------------
Mb3251|M.bovis_AF2122/97 --------------------------------------------------
Rv3224|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1333|M.marinum_M --------------------------------------------------
MUL_2546|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1894|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3946c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2386|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4262|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
MAV_1815|M.avium_104 --------------------------------------------------
TH_2687|M.thermoresistible__bu --------------------------------------------------
Mb3251|M.bovis_AF2122/97 --------------------------------------------------
Rv3224|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1333|M.marinum_M --------------------------------------------------
MUL_2546|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1894|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3946c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2386|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4262|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
MAV_1815|M.avium_104 --------------------------------------------------
TH_2687|M.thermoresistible__bu --------------------------------------------------
Mb3251|M.bovis_AF2122/97 --------------------------------------------------
Rv3224|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1333|M.marinum_M --------------------------------------------------
MUL_2546|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1894|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3946c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2386|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4262|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
MAV_1815|M.avium_104 --------------------------------------------------
TH_2687|M.thermoresistible__bu --------------------------------------------------
Mb3251|M.bovis_AF2122/97 --------------------------------------------------
Rv3224|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1333|M.marinum_M --------------------------------------------------
MUL_2546|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_1894|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3946c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2386|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4262|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
MAV_1815|M.avium_104 -----------------MGALSPRTLKDRVVVVTGASRGIGREIAIRAAA
TH_2687|M.thermoresistible__bu -----------------MNFQGGHTLAE--------TR------SLTDXA
Mb3251|M.bovis_AF2122/97 -----------------------MSLNGKTMFISGASRGIGLAIAKRAAR
Rv3224|M.tuberculosis_H37Rv -----------------------MSLNGKTMFISGASRGIGLAIAKRAAR
MMAR_1333|M.marinum_M -----------------------MSLNGKTMFISGASRGIGLAIAKRAAQ
MUL_2546|M.ulcerans_Agy99 -----------------------MSLNGKTMFISGASRGIGLAIAKRAAQ
MSMEG_1894|M.smegmatis_MC2_155 -----------------MVRMAQAPLSGKTMFISGASRGIGLAIAKRAAA
MAB_3946c|M.abscessus_ATCC_199 ----------------------MGSLAGKTIIMSGGSRGIGLAIALRAAR
Mflv_2386|M.gilvum_PYR-GCK -------------MASESGTSSTESLADRTLVVSGGSRGIGLAIALGAAR
Mvan_4262|M.vanbaalenii_PYR-1 ----------MSGTSNETGDIGGTRFAGRTMVVSGGSRGIGLAIALGAAR
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
: :
MAV_1815|M.avium_104 DGAAVALLAKTQTPNPKIAGTLTETAEAVRTAGGRALPLAVDVRDADGVA
TH_2687|M.thermoresistible__bu DGARVGLLARTETPNPKLAGTLAETAEAVRAAGGQAYEAVCDVRDADSVA
Mb3251|M.bovis_AF2122/97 DGANIALIAKTAEPHPKLPGTVFTAAKELEEAGGQALPIVGDIRDPDAVA
Rv3224|M.tuberculosis_H37Rv DGANIALIAKTAEPHPKLPGTVFTAAKELEEAGGQALPIVGDIRDPDAVA
MMAR_1333|M.marinum_M DGANIALIAKTAEPHPKLPGTVYTAAKELEEAGGQALPIVGDVRDPDSVS
MUL_2546|M.ulcerans_Agy99 DGANIALIAKTAEPHPKLPGTVYTAAKELEEAGGQALPIVGDVRDPDSVS
MSMEG_1894|M.smegmatis_MC2_155 DGANIALIAKTAEPHPKLEGTIYTAAEEIEDAGGQALPIVGDVRDGDSVA
MAB_3946c|M.abscessus_ATCC_199 DGANVAIMAKTAEPHPKLPGTIYTAAEEIEAAGGHALPLLGDVRDDEVVA
Mflv_2386|M.gilvum_PYR-GCK RGANVVLLAKTAEPHPRLPGTVHTAVADVEAAGGKGVAVVGDVRKEEDVA
Mvan_4262|M.vanbaalenii_PYR-1 QGANVVLLAKTAEPHPKLPGTVHTAVADVEAAGGKGVAVVGDVRKEEDVA
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAK-------NTAAQIVASGGVAYAYALDVSDAAAVE
** : : . :. : :** . *: . *
MAV_1815|M.avium_104 SAVAEAADEFGGIDVVVNNAGALDLRPTPKLPPKNFHRLLGVNVEGPFAV
TH_2687|M.thermoresistible__bu AAVADIADAFGGIDVVVNNAGALDLRPTSALPPKNFRRLLAVNVEGPFAV
Mb3251|M.bovis_AF2122/97 SAVATTVEQFGGIDICVNNASAINLGSITEVPMKRFDLMNGIQVRGTYAV
Rv3224|M.tuberculosis_H37Rv SAVATTVEQFGGIDICVNNASAINLGSITEVPMKRFDLMNGIQVRGTYAV
MMAR_1333|M.marinum_M AAVAKTVEQFGGIDICVNNASAINLGSITEVPMKRFDLMNGIQVRGTYAV
MUL_2546|M.ulcerans_Agy99 AAVAKTVEQFGGIDICVNNASAINLGSITEVPMKRFDLMNGIQVRGTYAV
MSMEG_1894|M.smegmatis_MC2_155 AAVTKAVEQFGGIDICVNNASAINLGSIEEVPLKRFDLMNGIQIRGTYAV
MAB_3946c|M.abscessus_ATCC_199 SAVAQTVEKFGGIDIVVNNASALNLAPSESISMKAYDLMQDINARGSFSL
Mflv_2386|M.gilvum_PYR-GCK RAIDTAVERFGGVDIVVNNASAIATEPTDALSAKKFDLMMDINIRGTFLL
Mvan_4262|M.vanbaalenii_PYR-1 RAIDTAVEHFGGVDVVVNNASAIATEPTEALSAKKFDLMMDINVRGTFLL
MLBr_00862|M.leprae_Br4923 AFAEQVSAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNC
.* *: ****. . : : : :: *
MAV_1815|M.avium_104 VQAAYTYLRNSDN-AHIVNISPPLNLDPRWVG-AHVGHTVGKYAESLLTL
TH_2687|M.thermoresistible__bu VQAALPHLRRSDN-AHIVNVSPPVNLAPAWIG-AHTGHTVGKYAESLLTI
Mb3251|M.bovis_AF2122/97 SQACIPHMKGREN-PHILTLSPPILLEKKWLR--PTAYMMAKYGMTLCAL
Rv3224|M.tuberculosis_H37Rv SQACIPHMKGREN-PHILTLSPPILLEKKWLR--PTAYMMAKYGMTLCAL
MMAR_1333|M.marinum_M SQACIPHLKGREN-PHILTLSPPVQLDKKWLK--PTAYMMAKFGMTLCAL
MUL_2546|M.ulcerans_Agy99 SQACIPHLKGREN-PHILTLSPPVQLDKKWLK--PTAYMMAKFGMTLCAL
MSMEG_1894|M.smegmatis_MC2_155 SQACIPHMKGREN-PHILTLSPPVRLEPEWLK--PTAYMMAKFGMTLCAL
MAB_3946c|M.abscessus_ATCC_199 STSAIGALKEADN-PHILTLSPPITLEPKWFERTTTAYTISKFSMSLVAI
Mflv_2386|M.gilvum_PYR-GCK TKAALPHLRQARAGAHVVTLAPPLNMNPHWLG-AHPSYTLSKYGMTLLSL
Mvan_4262|M.vanbaalenii_PYR-1 TKAALPHLRKSTSGAHVITLAPPLNMNPHWLG-AHPTYTLSKYGMTLLSL
MLBr_00862|M.leprae_Br4923 CRAFGQRLVERGTGGHIVNVSSMAAYAPLQSL---SAYCTSKAATYMFSD
: : *::.::. : .* . : :
MAV_1815|M.avium_104 GWAAEFASIPVAVNSLWPATTVASTGMIVAMGEDVVRAQARNTQIMADAV
TH_2687|M.thermoresistible__bu GWAAEFASIPVAVNSLWPATTVASTGMLVAMG-DEVRAQARDPRIMADAL
Mb3251|M.bovis_AF2122/97 GIAEEMRADGIASNTLWPRTMVATAAVQNLLGGDEAMARSRKPEVYADAA
Rv3224|M.tuberculosis_H37Rv GIAEEMRADGIASNTLWPRTMVATAAVQNLLGGDEAMARSRKPEVYADAA
MMAR_1333|M.marinum_M GIAEEMRDEGIASNTLWPRTLVATAAVQNLLGGDEAMGRARKPDVYADAA
MUL_2546|M.ulcerans_Agy99 GIAEEMRDEGIASNTLWPRTLVATAAVQNLLGGEEAMGRARKPDVYADAA
MSMEG_1894|M.smegmatis_MC2_155 GIAEEMREHGIASNTLWPRTLVATAAVQNLLGGDEAMARARKPQVYADAA
MAB_3946c|M.abscessus_ATCC_199 GLAAELRQYGIASNALWPRTTIDTAAIRNILG-AELVARCRSAQIMADAA
Mflv_2386|M.gilvum_PYR-GCK GWANEYADTGIGFSCLWPETYIATSAVTNLSDGDALVQASRSPDIMADAA
Mvan_4262|M.vanbaalenii_PYR-1 GWASEYADAGIGFSCLWPETYIATSAVSNLADGGDLVKSSRSPEIMGDAA
MLBr_00862|M.leprae_Br4923 CLRAELDAVGVGLTTICPG-VIDTNIIQSTRIDTPTAKQERVRDRRGQLD
* :. . : * : : : * .:
MAV_1815|M.avium_104 HALVTRPASDCSGHFYTDEEILREEGWDDDDLSEYLLAAREEDLTPNFYL
TH_2687|M.thermoresistible__bu HALVTRPA-DCTGNFYTDEQILREEGVAD--LTGYRLAASEDDLVPNFYL
Mb3251|M.bovis_AF2122/97 YVIVNKPATEYTGKTLLCEDVLVESGVTD--LSVYDCVPGAT-LGVDLWV
Rv3224|M.tuberculosis_H37Rv YVIVNKPATEYTGKTLLCEDVLVESGVTD--LSVYDCVPGAT-LGVDLWV
MMAR_1333|M.marinum_M YVILNKNSREYTGNSALCEDVLLESGVSD--LSIYNCVPGAK-LGVDLWV
MUL_2546|M.ulcerans_Agy99 YVILNKNSREYTGNSALCEDVLLESGVSD--LSIYNCVPGAK-LGVDLWV
MSMEG_1894|M.smegmatis_MC2_155 YAVFTKPAREFTGNTLLCEDVLLDSGVTD--LSVYDCVPGGE-LGVDLWV
MAB_3946c|M.abscessus_ATCC_199 YEILTKPSREVSGNCFIDDEVLREAGVTD--FSQYRECAEED-LELDFWM
Mflv_2386|M.gilvum_PYR-GCK VEILSRPAAEVNGQCFIDSEVLIASGVTD--LSRYG--GGDE-PILDIFV
Mvan_4262|M.vanbaalenii_PYR-1 VQILSRPPAEVNGQCYIDAAVLADSGVTD--LSGYG--GGDD-PILDIFV
MLBr_00862|M.leprae_Br4923 MMFRLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISR-VLPQVLR
. : . : * :.
MAV_1815|M.avium_104 SSTPLPSV---
TH_2687|M.thermoresistible__bu ESTPLPTI---
Mb3251|M.bovis_AF2122/97 EDANPPGYLPA
Rv3224|M.tuberculosis_H37Rv EDANPPGYLPA
MMAR_1333|M.marinum_M EEVNPPGYTQP
MUL_2546|M.ulcerans_Agy99 EEVNPPGYTQP
MSMEG_1894|M.smegmatis_MC2_155 DTPNPPGYTGP
MAB_3946c|M.abscessus_ATCC_199 ERA--------
Mflv_2386|M.gilvum_PYR-GCK DGRTS------
Mvan_4262|M.vanbaalenii_PYR-1 DGRVS------
MLBr_00862|M.leprae_Br4923 STARIRVI---
.