For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NLATQALAKAFERASNGLANPATLADPDRLRGAVTGKTALVTGASYGIGEATARKLAAAGATVLMVARSA ERLEDLVSAIAAGGGAAVAYPTDLTDEAAVGVLTKQVNQNHGPLDIVVSNAGKSLRRSLHDQYDRPHDFQ RTIDINYLGPIWLLLGLLPAMRESGRGHVVNVSSVGVRLVPGPQWGAYQASKGAFDRWLRSVAPELHGDG VDVTSVYFALVRTRMIEPTPMLGRLPGLEPDQAADAVAKAIVERPRTNEPPWLWPAELASVLLAGPAEQA ARVWHRRFFAPTAEAGKS*
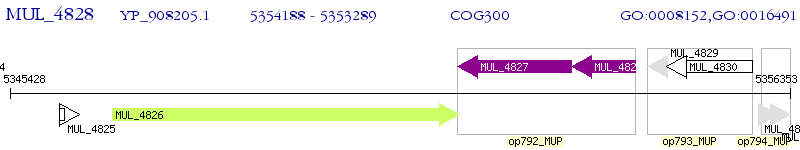
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4828 | - | - | 100% (299) | short-chain type dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_0918 | acrA1 | 2e-45 | 40.00% (250) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_0647 | - | 4e-44 | 41.26% (223) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1570 | - | 2e-56 | 46.95% (262) | fatty acyl-CoA reductase |
| M. gilvum PYR-GCK | Mflv_0026 | - | 3e-50 | 45.42% (240) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1543 | - | 2e-56 | 46.95% (262) | fatty acyl-CoA reductase |
| M. leprae Br4923 | MLBr_00862 | ephD | 6e-21 | 32.47% (231) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2540c | - | 5e-62 | 45.45% (275) | putative Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0295 | - | 1e-171 | 100.00% (299) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_5221 | - | 1e-136 | 77.85% (298) | NAD dependent epimerase/dehydratase family protein |
| M. smegmatis MC2 155 | MSMEG_1073 | - | 2e-50 | 46.85% (222) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3707 | - | 4e-26 | 36.41% (195) | PUTATIVE oxidoreductase, short-chain dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_0944 | - | 1e-49 | 45.42% (240) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4828|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0295|M.marinum_M --------------------------------------------------
MAV_5221|M.avium_104 --------------------------------------------------
MAB_2540c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1570|M.bovis_AF2122/97 --------------------------------------------------
Rv1543|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0944|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
MUL_4828|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0295|M.marinum_M --------------------------------------------------
MAV_5221|M.avium_104 --------------------------------------------------
MAB_2540c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1570|M.bovis_AF2122/97 --------------------------------------------------
Rv1543|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0944|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
MUL_4828|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0295|M.marinum_M --------------------------------------------------
MAV_5221|M.avium_104 --------------------------------------------------
MAB_2540c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1570|M.bovis_AF2122/97 --------------------------------------------------
Rv1543|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0944|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
MUL_4828|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0295|M.marinum_M --------------------------------------------------
MAV_5221|M.avium_104 --------------------------------------------------
MAB_2540c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1570|M.bovis_AF2122/97 --------------------------------------------------
Rv1543|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0944|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
MUL_4828|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0295|M.marinum_M --------------------------------------------------
MAV_5221|M.avium_104 --------------------------------------------------
MAB_2540c|M.abscessus_ATCC_199 --------------------------------------------------
Mb1570|M.bovis_AF2122/97 --------------------------------------------------
Rv1543|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_0026|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0944|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1073|M.smegmatis_MC2_155 --------------------------------------------------
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
MUL_4828|M.ulcerans_Agy99 -------------------------------------MNLATQALAKAFE
MMAR_0295|M.marinum_M -------------------------------------MNLATQALAKAFE
MAV_5221|M.avium_104 -------------------------MVQAARTNAAAKVNAIAQALDKVLN
MAB_2540c|M.abscessus_ATCC_199 --------------------------------MSTVLPSGPSPVVHHPLA
Mb1570|M.bovis_AF2122/97 -------------------------MNLGDLTNFVEKPLAAVSNIVNTPN
Rv1543|M.tuberculosis_H37Rv -------------------------MNLGDLTNFVEKPLAAVSNIVNTPN
Mflv_0026|M.gilvum_PYR-GCK ----------MRSSRKSASEVLLECEFGAGNRLRGVTSRNPLRRLADQVV
Mvan_0944|M.vanbaalenii_PYR-1 -----------------------------------MTSRNPLRRLADQVV
MSMEG_1073|M.smegmatis_MC2_155 ------------------------------------MSKHPLRRLTDNLF
TH_3707|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
MUL_4828|M.ulcerans_Agy99 RASNG----LANPATLADPDRLRGAVTGKTALVTGASYGIGEATARKLAA
MMAR_0295|M.marinum_M RASNG----LANPATLADPDRLRGAVTGKTALVTGASYGIGEATARKLAA
MAV_5221|M.avium_104 TVADR----VANPARVSDPDRLRGAVGGRTVLVTGASYGIGEATARRLAA
MAB_2540c|M.abscessus_ATCC_199 RAAAY----LIGPRGLRGTERLRSEVAGKIVVITGASYGLGAATAELFAQ
Mb1570|M.bovis_AF2122/97 SAGRYRPFYLRNLLDAVQGRNLNDAVKGKVVLITGGSSGIGAAAAKKIAE
Rv1543|M.tuberculosis_H37Rv SAGRYRPFYLRNLLDAVQGRNLNDAVKGKVVLITGGSSGIGAAAAKKIAE
Mflv_0026|M.gilvum_PYR-GCK LTSMRPP--ILPALLN-RPTRETIELRGKRVLLTGASSGIGEAAAEKLAR
Mvan_0944|M.vanbaalenii_PYR-1 LTSMRPP--ILPALLN-RPNRETIELKGKRVLLTGASSGIGEAAAEKLAR
MSMEG_1073|M.smegmatis_MC2_155 LASMRPP--VTDQLIQLRAARDAVDLTGKRVLITGASSGIGAAAARKFAE
TH_3707|M.thermoresistible__bu -----------------------VRADGR------ADQGLDAARX-----
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
. .. *:.
MUL_4828|M.ulcerans_Agy99 AGATVLMVARSAERLEDLV---SAIAAGGGAAVAYPTDLTDEAAVGVLTK
MMAR_0295|M.marinum_M AGATVLMVARSAERLEDLV---SAIAAGGGAAVAYPTDLTDEAAVGVLTK
MAV_5221|M.avium_104 AGATVLVVARSEERLGELT---AAINAGGGRAVAYPTDLTDESAVSALTK
MAB_2540c|M.abscessus_ATCC_199 AGAQVVLAARSLEQLRLVA---AGIAEKGGEAAVYRLDLTSEESITEFTE
Mb1570|M.bovis_AF2122/97 AGGTVVLVARTLENLENVANDIRAIRGNGGTAHVYPCDLSDMDAIAVMAD
Rv1543|M.tuberculosis_H37Rv AGGTVVLVARTLENLENVANDIRAIRGNGGTAHVYPCDLSDMDAIAVMAD
Mflv_0026|M.gilvum_PYR-GCK RGATVVAVARRQDLLDELV---TRIADAGGDARAHACDLSDLDAVDELVA
Mvan_0944|M.vanbaalenii_PYR-1 RGATVVAVARRGDLLDDLV---ARITDAGGRARAHACDLSDLDAIDALVA
MSMEG_1073|M.smegmatis_MC2_155 RGARVIVVARRAELLDEVA---GGITASGGDAKARPCDLTDLDAIDALVA
TH_3707|M.thermoresistible__bu ----------------------------GGTGRALACDLTDLDAIDELVL
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTA---AQIVASGGVAYAYALDVSDAAAVEAFAE
** . . *::. :: :.
MUL_4828|M.ulcerans_Agy99 QVNQNHGPLDIVVSNAGKSLRRSLHDQYDRPHDFQRTIDINYLGPIWLLL
MMAR_0295|M.marinum_M QVNQNHGPLDIVVSNAGKSLRRSLHDQYDRPHDFQRTIDINYLGPIWLLL
MAV_5221|M.avium_104 QITEEHGPLDVVVSNAGKSLRRSLHHQYDRPHDFQRTIDVNYLGPVRLLL
MAB_2540c|M.abscessus_ATCC_199 AVLHDFGEVDYLIHNAGKSLRRSIHLSYDRPKDVNATSGANYVGPMRLTL
Mb1570|M.bovis_AF2122/97 QVLGDLGGVDILINNAGRSIRRSLELSYDRIHDYQRTMQLNYLGAVQLIL
Rv1543|M.tuberculosis_H37Rv QVLGDLGGVDILINNAGRSIRRSLELSYDRIHDYQRTMQLNYLGAVQLIL
Mflv_0026|M.gilvum_PYR-GCK TVEKDLGGIDILVNNAGRSIRRPLEESLERWHDVERTMTLNYYGPLRLIR
Mvan_0944|M.vanbaalenii_PYR-1 TVEDELGGVDILVNNAGRSIRRPLEESLDRWHDVERTMTLNYYGPLRLIR
MSMEG_1073|M.smegmatis_MC2_155 EIESDHGGLDILVNNAGHSIRRPLADSLERWHDIERTMTLNYYSPLRLIR
TH_3707|M.thermoresistible__bu KIEQDLGGVDILINNAGKSIRRPLAESLERWHDVERTMNLNYYAPLRLIR
MLBr_00862|M.leprae_Br4923 QVSAKHGVPDIVVNNAGIGQAGRFLDTP--PEEFDRVLAVNLGGVVNCCR
: . * * :: *** . : .: : . * . :
MUL_4828|M.ulcerans_Agy99 GLLPAMRESGRGHVVNVSSVGVRLVPGPQWGAYQASKGAFDRWLRSVAPE
MMAR_0295|M.marinum_M GLLPAMRESGRGHVVNVSSVGVRLVPGPQWGAYQASKGAFDRWLRSVAPE
MAV_5221|M.avium_104 GLLPAMRDNGRGHVVNVSSVGVRVVPGPQWGAYQASKGAFDRWLRSVAPE
MAB_2540c|M.abscessus_ATCC_199 ALLPGMRARGSGHIVNVSTVGVMFGVVPKWGFYLSSKAAFDIWMRAVGME
Mb1570|M.bovis_AF2122/97 KFIPGMRERHFGHIVNVSSVGVQTR-APRFGAYIASKAALDSLCDALQAE
Rv1543|M.tuberculosis_H37Rv KFIPGMRERHFGHIVNVSSVGVQTR-APRFGAYIASKAALDSLCDALQAE
Mflv_0026|M.gilvum_PYR-GCK GLAPGMLARGDGHIINVSTWGVKTESPPLFGVYNASKAALSSVSRIIETE
Mvan_0944|M.vanbaalenii_PYR-1 GLAPGMLERGDGHVINVSTWGVKTESPPLFGVYNASKAALSSISRIMETE
MSMEG_1073|M.smegmatis_MC2_155 GFAPGMRERGAGHIINVSTWGVLSEASPLFSVYQASKAALSAVSRVIETE
TH_3707|M.thermoresistible__bu GLAPGMRE----------TWGVLSEASPLFSVYNASKAALTAVSRVVETE
MLBr_00862|M.leprae_Br4923 AFGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAE
: : : . . * :**.* : *
MUL_4828|M.ulcerans_Agy99 LHGDGVDVTSVYFALVRTRMIEPTPMLGRLPGLEPDQ----AADAVAKAI
MMAR_0295|M.marinum_M LHGDGVDVTSVYFALVRTRMIEPTPMLGRLPGLEPDQ----AADAVAKAI
MAV_5221|M.avium_104 LHADGVHVSTVYFALVRTRMIAPTPVLGRLPGLSPDQ----AADVIAKAV
MAB_2540c|M.abscessus_ATCC_199 ARGDGVTLTTFYAGLMHTRMSAPSRWMRLLPGQTPLD----AAKVLARAV
Mb1570|M.bovis_AF2122/97 TVHDNVRFTTVHMALVRTPMISPTTIYDKFPTLTPDQ----AAGVITDAI
Rv1543|M.tuberculosis_H37Rv TVHDNVRFTTVHMALVRTPMISPTTIYDKFPTLTPDQ----AAGVITDAI
Mflv_0026|M.gilvum_PYR-GCK WSGRGVHATTLYYPLVKTPMIAPTRAYDGLPGLSADE----AAGWIVTAA
Mvan_0944|M.vanbaalenii_PYR-1 WADRGVHSTTLYYPLVKTPMIAPTRAYDGLPGLSAEE----AAGWIITAA
MSMEG_1073|M.smegmatis_MC2_155 WGDGGVHSTTLYYPLVDTPMIKPTRAFDGLPALSADE----AADWMITAA
TH_3707|M.thermoresistible__bu WSQFGVHSTTLFYPLVKTPMIAPTREYDNLPALSAEE----AADWMISAA
MLBr_00862|M.leprae_Br4923 LDAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLR
.* ::. :: * : .: . :
MUL_4828|M.ulcerans_Agy99 VERPRTNEPPWLWPAELASVLLAGPAEQAARVWHRRFFAPTAEAGKS---
MMAR_0295|M.marinum_M VERPRTNEPPWLWPAELASVLLAGPAEQAARVWHRRFFAPTAEAGKS---
MAV_5221|M.avium_104 IERPRTLEPPWVLPAELASVLLAGPADRAARLWHRRFFTDAEEAGRER--
MAB_2540c|M.abscessus_ATCC_199 VDKPRTLTPAYGHPFAILAPVLRFPLEPILGFVYRRLGDTQASLARVTAG
Mb1570|M.bovis_AF2122/97 VHRPRRASSPFGQFAAVADAVNPAVMDRVRNRAFNMFGDSSAAKGSES--
Rv1543|M.tuberculosis_H37Rv VHRPRRASSPFGQFAAVADAVNPAVMDRVRNRAFNMFGDSSAAKGSES--
Mflv_0026|M.gilvum_PYR-GCK RHRPVSIAPRIAVTAHAINSVAPGAVDTIMKRQRMQPRD-----------
Mvan_0944|M.vanbaalenii_PYR-1 RHRPVSIAPRIAVTAHAINSIAPAAVDTIMKRQRMQPKD-----------
MSMEG_1073|M.smegmatis_MC2_155 RHRPVQIAPRVAVGARALNAVAPGVVNAVMKRQSAQPDGVGRRPIFPAG-
TH_3707|M.thermoresistible__bu RYRPIQIAPRMAVTARALNVVAPGVVNAVMKRQRIQP-------VFV---
MLBr_00862|M.leprae_Br4923 RYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIRV
* : :
MUL_4828|M.ulcerans_Agy99 -----------------------
MMAR_0295|M.marinum_M -----------------------
MAV_5221|M.avium_104 -----------------------
MAB_2540c|M.abscessus_ATCC_199 QSTTTRIDAPERIGVHAE-----
Mb1570|M.bovis_AF2122/97 QTDTSELDKRSETFVRATRGIHW
Rv1543|M.tuberculosis_H37Rv QTDTSELDKRSETFVRATRGIHW
Mflv_0026|M.gilvum_PYR-GCK -----------------------
Mvan_0944|M.vanbaalenii_PYR-1 -----------------------
MSMEG_1073|M.smegmatis_MC2_155 -----------------------
TH_3707|M.thermoresistible__bu -----------------------
MLBr_00862|M.leprae_Br4923 I----------------------