For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSQDLTGRTAIITGASRGIGLAIAQQLAAAGANVVLTARKQEAADEAAAQVGPQALGVGAHAVDEDAARQ CVQLTLERFGSVDILVNNAGTNPAYGPLIEQDHARFAKIFDVNLWAPLLWTSLAVKSWMGEHGGSIVNTA SIGGLHQSPAMGMYNATKAALIHVTKQLALELSPRVRVNAIAPGVVRTRLAEALWKDHEDPLSSSIALGR IGEPIDVAAAVAFLVSDAASWITGETLVIDGGTVLGAAQGFRSAPTPS*
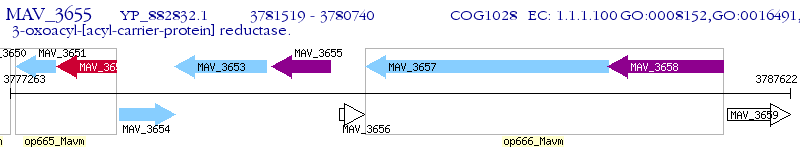
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3655 | fabG | - | 100% (259) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. avium 104 | MAV_2775 | - | 1e-35 | 37.25% (247) | short chain dehydrogenase |
| M. avium 104 | MAV_1572 | - | 6e-35 | 36.93% (241) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2788c | fabG | 1e-130 | 87.55% (257) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1202 | - | 1e-38 | 38.11% (244) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2766c | fabG | 1e-130 | 87.55% (257) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 2e-30 | 32.92% (240) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0833 | - | 2e-41 | 39.59% (245) | Short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_1952 | fabG | 1e-126 | 85.55% (256) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_1740 | - | 3e-40 | 36.21% (243) | dehydrogenase/reductase SDR family protein member 4 |
| M. thermoresistible (build 8) | TH_2589 | - | 2e-41 | 38.68% (243) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_2173 | fabG | 1e-125 | 85.16% (256) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_5605 | - | 2e-37 | 37.70% (244) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1202|M.gilvum_PYR-GCK -----MT---GFADRLFDLTDRVVLITGGSRGLGREMAVAAAHCGADVII
Mvan_5605|M.vanbaalenii_PYR-1 -----MTASGGFADQLFDLTDRVVLVTGGSRGLGREMAFAAARCGADVVI
MSMEG_1740|M.smegmatis_MC2_155 -----MDR--QRFERLFDMSDRTVIVTGGTRGIGLSLAEGFVLAGARVVV
TH_2589|M.thermoresistible__bu -----VDR--QTFDKLFDMTGRTVVVTGGTRGIGLALAEGYVLAGARVVV
MAB_0833|M.abscessus_ATCC_1997 -----MNR--ETFSRLFDLTGRTAIVTGGTRGIGRALAEGYACAGANVVV
MMAR_1952|M.marinum_M -----MTS--------LDLTGRTAIITGASRGIGLAIAQRLAGAGANVVL
MUL_2173|M.ulcerans_Agy99 -----MTS--------LDLTGRTAIITGASRGIGLAIAQRLAGAGANVVL
Mb2788c|M.bovis_AF2122/97 -----MTS--------LDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVL
Rv2766c|M.tuberculosis_H37Rv -----MTS--------LDLTGRTAIITGASRGIGLAIAQQLAAAGAHVVL
MAV_3655|M.avium_104 -----MTS--------QDLTGRTAIITGASRGIGLAIAQQLAAAGANVVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
: .* ::**..**:* :* . * * :
Mflv_1202|M.gilvum_PYR-GCK ASRKYDACVATAEEITAQTGRAALPYGVHVGRWDELDGLVDAAYARFGKV
Mvan_5605|M.vanbaalenii_PYR-1 ASRKYEACVATADEIAAETGRAAFPYQVHVGRWDELDGLVDAAYDRFGRV
MSMEG_1740|M.smegmatis_MC2_155 ASRKPDACETAAAHLRELGG-DAVGVPAHTGDVDDLGAVVERAVSEFGGV
TH_2589|M.thermoresistible__bu ASRKSDACERAAAHLRALGG-DAIGVPTHLGDIDACEALVRRTVDEFGGI
MAB_0833|M.abscessus_ATCC_1997 ASRKPEACTEAVERLRSLGA-RALGVPTHTGDIHSLEELVRATAEEFGGI
MMAR_1952|M.marinum_M TARKQESADAAAAEV----GERAVGVGAHAVDEEAARGCVDLALERFGSV
MUL_2173|M.ulcerans_Agy99 TARKQESADAAAAEV----GERAVGVGAHAVDEEAARGCVDLALERFGSV
Mb2788c|M.bovis_AF2122/97 TARRQEAADEAAAQV----GDRALGVGAHAVDEDAARRCVDLTLERFGSV
Rv2766c|M.tuberculosis_H37Rv TARRQEAADEAAAQV----GDRALGVGAHAVDEDAARRCVDLTLERFGSV
MAV_3655|M.avium_104 TARKQEAADEAAAQV----GPQALGVGAHAVDEDAARQCVQLTLERFGSV
MLBr_01807|M.leprae_Br4923 THRASVVPD------------WFFGVECDVTDNDAIGAAFKAVEEHQGPV
: * . . . . . . * :
Mflv_1202|M.gilvum_PYR-GCK DVLVNNAGMSPVYDKQSDVTEKLFDAVVNLNLKGPFRLSALIGERMVADG
Mvan_5605|M.vanbaalenii_PYR-1 DVLVNNAGMSPLYESLSAVTEKLFDSVVNLNLKGPFRLSVLVGDRMVADG
MSMEG_1740|M.smegmatis_MC2_155 DVVVNNAAN-ALAQPLGKMTPEAWAKSYEVNLRGPVFLVQHALPYLQKSQ
TH_2589|M.thermoresistible__bu DVLVNNAAN-ALTQPLGELTVDAWTKSFEVNLRGPVFLVQAALPHLKTSP
MAB_0833|M.abscessus_ATCC_1997 DIVVNNAAN-ALTEPVGAFTVDGWDKSFGVNLRGPVFLVQAALPHLVASP
MMAR_1952|M.marinum_M DILINNAGTNPAYGPLIEQDHARFTKIFDVNLWAPLMWTSLVVKAWMGEH
MUL_2173|M.ulcerans_Agy99 DILINNAGTNPAYGPLIEQDHARFTKIFDVNLWAPLMWTSLVVKAWMGEH
Mb2788c|M.bovis_AF2122/97 DILINNAGTNPAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEH
Rv2766c|M.tuberculosis_H37Rv DILINNAGTNPAYGPLLEQDHARFAKIFDVNLWAPLMWTSLVVTAWMGEH
MAV_3655|M.avium_104 DILVNNAGTNPAYGPLIEQDHARFAKIFDVNLWAPLLWTSLAVKSWMGEH
MLBr_01807|M.leprae_Br4923 EVLVANAGITSDMLIMR-MSEEQFQKVIDVNLTGVFRVAKLASRNMIKQR
:::: **. . : :** . . .
Mflv_1202|M.gilvum_PYR-GCK GGSIVNVSTHGSLRPHPSFIPYAASKAGLNAMTEGLALAFGPS-VRVNTL
Mvan_5605|M.vanbaalenii_PYR-1 GGSIINVSTHGSIRPHPSFVPYAASKAGLNAMTEALALAYGPT-VRVNTL
MSMEG_1740|M.smegmatis_MC2_155 NAAVLNMVSVGAFNFAPALSIYASSKAALMSVTRSMAAEFAPSGIRVNAL
TH_2589|M.thermoresistible__bu HAAVLNMVSVGAFLFSPGTAMYSAGKAALMSFTRSMAAQFAPLGIRVNAL
MAB_0833|M.abscessus_ATCC_1997 HAAVLNVVSVGAFMFGQGVSMYSAAKAALVSYTRSAAAELASRGVRVNAL
MMAR_1952|M.marinum_M GGVVINTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-VRVNAI
MUL_2173|M.ulcerans_Agy99 GGVVINTASIGGMHQSPAMGMYNATNAALIHVTKQLALELSPR-VRVNAI
Mb2788c|M.bovis_AF2122/97 GGAVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVNAI
Rv2766c|M.tuberculosis_H37Rv GGAVVNTASIGGMHQSPAMGMYNATKAALIHVTKQLALELSPR-IRVNAI
MAV_3655|M.avium_104 GGSIVNTASIGGLHQSPAMGMYNATKAALIHVTKQLALELSPR-VRVNAI
MLBr_01807|M.leprae_Br4923 FGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITANVV
. : : .: . * : ::.* :. * .. : .*.:
Mflv_1202|M.gilvum_PYR-GCK MPGPFLTDISNAWNF-EGTDNPFRGSALQRAGQPAEIVGAALFLMSDASS
Mvan_5605|M.vanbaalenii_PYR-1 MPGPFLTDISDAWTFTDGDTNPFGHSALQRAGQPAEIVGAALFLMSDASS
MSMEG_1740|M.smegmatis_MC2_155 APGPVDTDMMRKNPQ-EAIDAMTAGTLMKRLAHPDEMVGAALLLCSDAGS
TH_2589|M.thermoresistible__bu APGPVDTDMMRNNPQ-EVVDMMAGATLMKRLATPDEMVGAALLLTSDAGS
MAB_0833|M.abscessus_ATCC_1997 APGAIDTDMVRKTPP-AEVERMANTNHLKRLGLPDEMVGTALLLTSDAGS
MMAR_1952|M.marinum_M CPGVVRTKLAEALWK-DHEDPLAANIALGRIGEPVDVAEAVGFLVSDAAS
MUL_2173|M.ulcerans_Agy99 CPGVVRTKLAEALWK-DHEDPLAANIALGRIGEPVDVAEAVGFLVSDAAS
Mb2788c|M.bovis_AF2122/97 CPGVVRTRLAEALWK-DHEDPLAATIALGRIGEPADIASAVAFLVSDAAS
Rv2766c|M.tuberculosis_H37Rv CPGVVRTRLAEALWK-DHEDPLAATIALGRIGEPADIASAVAFLVSDAAS
MAV_3655|M.avium_104 APGVVRTRLAEALWK-DHEDPLSSSIALGRIGEPIDVAAAVAFLVSDAAS
MLBr_01807|M.leprae_Br4923 APGFIDTEMTRAMDS-KYQEAALQAIPLGRIGAVEEIAGAVSFLASEDAG
** . * : : * . ::. :. :* *: ..
Mflv_1202|M.gilvum_PYR-GCK FTTGSIVRADGGSA---------------
Mvan_5605|M.vanbaalenii_PYR-1 FTTGSIVRADGGIP---------------
MSMEG_1740|M.smegmatis_MC2_155 YMTGQVVIVDGGGTPR-------------
TH_2589|M.thermoresistible__bu YITGTVLIADGGGTPR-------------
MAB_0833|M.abscessus_ATCC_1997 YITGQTIIADGGLVAR-------------
MMAR_1952|M.marinum_M WITGETMVIDGGLLLGSAAGFQSRPGSAS
MUL_2173|M.ulcerans_Agy99 WITGETMVIDGGLLLGSAAGFQSRPGSAS
Mb2788c|M.bovis_AF2122/97 WITGETMIIDGGLLLGNALGFRAAPSTEH
Rv2766c|M.tuberculosis_H37Rv WITGETMIIDGGLLLGNALGFRAAPSTEH
MAV_3655|M.avium_104 WITGETLVIDGGTVLGAAQGFRSAPTPS-
MLBr_01807|M.leprae_Br4923 YISGAVIPVDGGLGMGH------------
: :* : ***