For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PEYDYEELKKEAEQYIKFEKDKKNRIAYITFDRPDAQNSATLGMRQNYADLIHKCNVDDDVKVVVIRGEG EDFGSGGDLPEQRGMLENPGMPLLHELAINDDDVKYPPGGSYRYLSTVTDFYAKARAGNRPLQELRKISI IEAKGYCYGWHFYQAGDADLVIASDDALFGHPAFRYVGWGPRLWWWAETMGLRKFSEMLFTGRPFSAKEM YDCGFVNSVVPRDKLEAETEKYALACSKARPTDTVAVQKTFLELYKQHKGEYFGSLLTGLVEGMLPLIAN DAEEVDLTAGTFEKGLNNVVKDNDLNFPPEWRLSRSGRAKP*
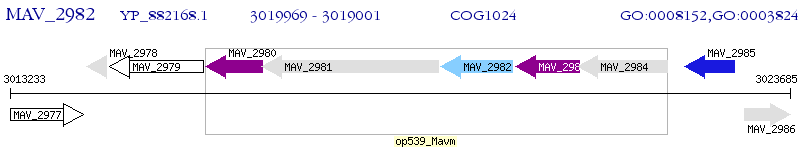
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2982 | - | - | 100% (322) | enoyl-CoA hydratase/isomerase |
| M. avium 104 | MAV_0670 | - | 9e-18 | 28.09% (235) | enoyl-CoA hydratase |
| M. avium 104 | MAV_4694 | - | 1e-16 | 26.76% (299) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0464c | echA2 | 3e-17 | 27.09% (299) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2422 | - | 1e-101 | 56.91% (311) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv0456c | echA2 | 3e-17 | 27.09% (299) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 1e-10 | 25.89% (224) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_3859c | - | 5e-17 | 26.39% (216) | enoyl-CoA hydratase |
| M. marinum M | MMAR_4758 | echA4_1 | 1e-107 | 58.58% (309) | enoyl-CoA hydratase, EchA4_1 |
| M. smegmatis MC2 155 | MSMEG_4852 | - | 1e-102 | 56.41% (312) | enoyl-CoA hydratase, putative |
| M. thermoresistible (build 8) | TH_2025 | - | 1e-104 | 58.01% (312) | enoyl-CoA hydratase, putative |
| M. ulcerans Agy99 | MUL_1414 | echA2 | 4e-16 | 25.93% (297) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4230 | - | 1e-101 | 56.59% (311) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0464c|M.bovis_AF2122/97 ------------MPTPDFQTLLYTTAGPVATITLNRPEQLNTIVPPMPDE
Rv0456c|M.tuberculosis_H37Rv ------------MPTPDFQTLLYTTAGPVATITLNRPEQLNTIVPPMPDE
MUL_1414|M.ulcerans_Agy99 ------------MPTPEFQTLLYSTAGPVATITLNRPEHLNTIVPPMPDE
MAB_3859c|M.abscessus_ATCC_199 -------------MVDTLKTMTYEVTDRVARITFNRPEQGNAIIADTPLE
Mflv_2422|M.gilvum_PYR-GCK MAERPSPSAAERPTPEEIILYEKDPQTKIATITFNRPEFLNAPTSMARLR
Mvan_4230|M.vanbaalenii_PYR-1 MAERPQPPAAERPAPEEIILYDKDPKTKIATITFNRPEFLNAPTSMARLR
MSMEG_4852|M.smegmatis_MC2_155 --------MAERPTPEEIILYDKDPETKIATITFNRPEFLNAPTSMARLR
TH_2025|M.thermoresistible__bu --------MADRPAPEEIILYDKDPRTKIATITFNRPEFLNAPTSAARLR
MMAR_4758|M.marinum_M -------MPKSEPPVDKVIRYDKDPETRIATITFDRPGHLNAPTIAARLR
MAV_2982|M.avium_104 --MPEYDYEELKKEAEQYIKFEKDKKNRIAYITFDRPDAQNSATLGMRQN
MLBr_02402|M.leprae_Br4923 ---------------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNE
:. **::** *: .
Mb0464c|M.bovis_AF2122/97 IEAAIGLAERDQDIKVIVLRGAGRAFSGGYDFG--------GGFQHWGDA
Rv0456c|M.tuberculosis_H37Rv IEAAIGLAERDQDIKVIVLRGAGRAFSGGYDFG--------GGFQHWGDA
MUL_1414|M.ulcerans_Agy99 IEAAVGLAERDQGIKVIVLRGAGRAFSGGYDFG--------GGFQHWGET
MAB_3859c|M.abscessus_ATCC_199 LAALVERADLDPQVHVILVSGRGDGFCGGFDLSAYAEANDEGSVRYSGTA
Mflv_2422|M.gilvum_PYR-GCK YADVLRAANADNDVKVVIIRGVGDNLGSGADLPEFMEGNDNP--AVRLAE
Mvan_4230|M.vanbaalenii_PYR-1 YADVLRAANTDNDVKVVIIRGVGDNLGSGADLPEFMEGNDNP--AARLAE
MSMEG_4852|M.smegmatis_MC2_155 YADVLRAANADNDVKVVVIRGVGDNLGSGADLPEFMEGNDNP--AVRLAE
TH_2025|M.thermoresistible__bu YADLLRAATVDNDVKVVVIRGVGDNLGSGADLPEFMEGNDNP--ALRLAE
MMAR_4758|M.marinum_M YADLLHRASIDDEVKVLVIRGEGEDLGSGADLEEFMEVMNSPDQGPRLAE
MAV_2982|M.avium_104 YADLIHKCNVDDDVKVVVIRGEGEDFGSGGDLPEQRGMLENPG-MPLLHE
MLBr_02402|M.leprae_Br4923 ITNAAKELDIDPDVGAILITGSPKVFAAGADIK-----------------
* : .::: * : .* *:
Mb0464c|M.bovis_AF2122/97 M---------MTDGRWDPGKDFAMVTARETGPTQKFMAIWRASKPVIAQV
Rv0456c|M.tuberculosis_H37Rv M---------MTDGRWDPGKDFAMVTARETGPTQKFMAIWRASKPVIAQV
MUL_1414|M.ulcerans_Agy99 M---------MTDGQWDPGKDFAMVTARETAPTQKFMAIWRASKPVIAQI
MAB_3859c|M.abscessus_ATCC_199 LDGQVQLRNHLPNINWDPMLDYQMMSR----FTRGFSSLLHANKPTVVKV
Mflv_2422|M.gilvum_PYR-GCK LRLEDDGVGEVTYPPKGTFRNGATISAWYANSQAGNRALQDFKKISIVEA
Mvan_4230|M.vanbaalenii_PYR-1 LRLEDDGVGEVTYPPKGTFRNGATISAWYANSQAGNRALQDFKKISIVEA
MSMEG_4852|M.smegmatis_MC2_155 LRLEDDGVGEVTYPPRGTFRNGATISAWYANSQAGNRALQDFKKISIVEA
TH_2025|M.thermoresistible__bu LRLEDDGVGEVTYPPKGTFRNGATISAWYANSQAGNRPLQELKKISIVEA
MMAR_4758|M.marinum_M FRIGPE---EVKYPPLGSFRHGASVSQWYANPNAGIRGLQDFKKISILEV
MAV_2982|M.avium_104 LAINDD---DVKYPPGGSYRYLSTVTDFYAKARAGNRPLQELRKISIIEA
MLBr_02402|M.leprae_Br4923 -----------------EMASLTFTDAFDADFFSAWGKLAAVRTPMIAAV
: . :
Mb0464c|M.bovis_AF2122/97 HGWCVGGASDYALCADIVIASEDAVIGTPYSRMWG-AYLTG--MWLYRLS
Rv0456c|M.tuberculosis_H37Rv HGWCVGGASDYALCADIVIASEDAVIGTPYSRMWG-AYLTG--MWLYRLS
MUL_1414|M.ulcerans_Agy99 HGWCVGGASDYALCADIVIASDDAVIGTPYSRMWG-AYLSG--MWLYRLS
MAB_3859c|M.abscessus_ATCC_199 HGYCVAGGTDIALHADQLIVAADAKIGYPPTRVWG-VPATG--LWAHKLG
Mflv_2422|M.gilvum_PYR-GCK KGYCYGWHFYQCADADLVISSDDALFGHPSFRYHGWGPRMW--TWVQMMG
Mvan_4230|M.vanbaalenii_PYR-1 KGYCYGWHFYQCADADLVISSDDALFGHPSFRYHGWGPRMW--TWVQMMG
MSMEG_4852|M.smegmatis_MC2_155 KGYCYGWHFYQCADADLVISSDDALFGHPSFRYHGWGPRMW--TWVQMMG
TH_2025|M.thermoresistible__bu KGYCYGWHFYQCADADLVISSDDALFGHPSFRYYGWGPRMW--TWVQMMG
MMAR_4758|M.marinum_M KGYCYGWHFYQAADADLVISSDDALFGHPSFRYYGWGPRMW--WWAQTMG
MAV_2982|M.avium_104 KGYCYGWHFYQAGDADLVIASDDALFGHPAFRYVGWGPRLW--WWAETMG
MLBr_02402|M.leprae_Br4923 AGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIG
*:. . . .* :* : * :* * : :.
Mb0464c|M.bovis_AF2122/97 LAKVKWHSLTGRPLTGVQAAEAELINEAVPFERLEARVAEIATELAR-IP
Rv0456c|M.tuberculosis_H37Rv LAKVKWHSLTGRPLTGVQAAEAELINEAVPFERLEARVAEIATELAR-IP
MUL_1414|M.ulcerans_Agy99 FAKAKWHSLTGRPLSGVQAAQIELINEAVPFERLEARVAEVAAELAQ-IP
MAB_3859c|M.abscessus_ATCC_199 DQRAKRLLLTGDCLSGRQAYDWGLAVEAPEPDELDERTERLVERIAA-MP
Mflv_2422|M.gilvum_PYR-GCK LRKFQEMVFTGRPFTAAEMYDCNFLNKVVERDQLEAETQKYALACARNRP
Mvan_4230|M.vanbaalenii_PYR-1 LRKFQEMVFTGRPFTAAEMYDCNFLNKVVTRDQLEAETRKYALACARNRP
MSMEG_4852|M.smegmatis_MC2_155 LRKFQEMVFTGRPFTAAEMYDCNFLNKVVPRDRLEAEVDKYATACARNRP
TH_2025|M.thermoresistible__bu LRKFSEMVFTGRPFTAAEMYDCNFLNKVVPRDQLEAEVDKYARACARNRP
MMAR_4758|M.marinum_M IRKFQEMVFTGRAFTAAEMQDCNFLNSVVPREELEAEVAKYALACARNRP
MAV_2982|M.avium_104 LRKFSEMLFTGRPFSAKEMYDCGFVNSVVPRDKLEAETEKYALACSKARP
MLBr_02402|M.leprae_Br4923 KAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVATTISQMSR
: :** : . : : . : * .. . :
Mb0464c|M.bovis_AF2122/97 LSQLQAQKLIVNQAYENMGLASTQLLGGILDGLMR---NTPDALEFIRTA
Rv0456c|M.tuberculosis_H37Rv LSQLQAQKLIVNQAYENMGLASTQLLGGILDGLMR---NTPDALEFIRTA
MUL_1414|M.ulcerans_Agy99 LSQLQAQKLIVNQAYENMGLASTQTLGGILDGLMR---NTPDALDFIKTA
MAB_3859c|M.abscessus_ATCC_199 LNQLMMIKLACNSVLINQGIANSAMIGTVFDGISR---HTPEAHAFTADA
Mflv_2422|M.gilvum_PYR-GCK VDTVFQQKMFFEIFKQQQGEYMGSLLSAFFESMGNGVANDSDEDLDMFES
Mvan_4230|M.vanbaalenii_PYR-1 VDTVFQQKMFFEIFKQQQGEYMGSLLSAFFESMGNGVANDSDDDLDMFES
MSMEG_4852|M.smegmatis_MC2_155 VDTVFQQKMFFEIFKQQQGEYMGSLLSAFFESMGNGVANDSDDDLDMFES
TH_2025|M.thermoresistible__bu VDTVFQQKIFFEVFKQHQGEYMGSLLSAFFESMGSGAVNDDTDDLDMHEA
MMAR_4758|M.marinum_M NDTVFMQKVFFEIMKQFQGEYMGSLLSGFFESMGSGVRADT-DDLMLDDA
MAV_2982|M.avium_104 TDTVAVQKTFLELYKQHKGEYFGSLLTGLVEGMLPLIANDAEEVDLTAGT
MLBr_02402|M.leprae_Br4923 SATRMAKEAVNRSFESTLAEGLLHERRLFHS-------------TFVTDD
: . . . .
Mb0464c|M.bovis_AF2122/97 QTQGVRAAVERRDGPFGDYSQAPPELRPDPTHVITPDGSM---
Rv0456c|M.tuberculosis_H37Rv QTQGVRAAVERRDGPFGDYSQAPPELRPDPTHVITPDGSM---
MUL_1414|M.ulcerans_Agy99 ENQGVRAAVERRDGPFGDYSQAPPELRPNPNHVIVPDRDR---
MAB_3859c|M.abscessus_ATCC_199 IANGYRQAVRHRDEPFGDYGRKPTEL-----------------
Mflv_2422|M.gilvum_PYR-GCK IDSGLSAAVNDNDSKFPPEFRLSKRNRAKD-------------
Mvan_4230|M.vanbaalenii_PYR-1 IDSGLSAAVNDNDSKFPPEFRLSKRNRAKKD------------
MSMEG_4852|M.smegmatis_MC2_155 IDSGLSAAVNDNDSRFPPEFRLSKKNRAKP-------------
TH_2025|M.thermoresistible__bu IDSGLADAVNDNDMRFPPEFRLSKSNRAKE-------------
MMAR_4758|M.marinum_M IEQGLGDAVKDNDSRFPPEWRLSKKSRAKDGTGKPAKQKKKKG
MAV_2982|M.avium_104 FEKGLNNVVKDNDLNFPPEWRLSRSGRAKP-------------
MLBr_02402|M.leprae_Br4923 QSEGMAAFIEKRAPQFTHR------------------------
.* :. . *