For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPSDFETLLYTTAGPVATITLNRPDQLNTIVPPMPDEIEAAVGRAERDPAIKVIVLRGAGRAFSGGYDFG GGFQHWGEAMMTDGRWDPGKDFAMVSARETGPTQKFMAIWRASKPVIAQVHGWCVGGASDYALCADLVIA SEDAVIGTPYSRMWGAYLTGMWLYRLSLAKAKWHALTGRPLTGVQAAEIELINEAVPFERLEARVAEVAA ELARIPLSQLQAQKLIVNQAYENMGLASTQTLGGILDGLMRNTPDALEFIKIAETRGVRAAVERRDGPFG DYSQAPPELRPDPSHVIVPDRDG
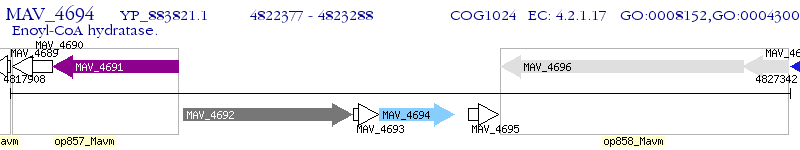
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4694 | - | - | 100% (303) | enoyl-CoA hydratase |
| M. avium 104 | MAV_4499 | - | 3e-48 | 37.05% (305) | enoyl-CoA hydratase |
| M. avium 104 | MAV_2950 | - | 7e-32 | 30.45% (266) | enoyl-CoA hydratase/isomerase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0464c | echA2 | 1e-164 | 92.36% (301) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2433 | - | 5e-23 | 32.02% (228) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv0456c | echA2 | 1e-164 | 92.36% (301) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 4e-15 | 25.09% (283) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_3859c | - | 1e-51 | 37.99% (308) | enoyl-CoA hydratase |
| M. marinum M | MMAR_0778 | echA2 | 1e-162 | 90.43% (303) | enoyl-CoA hydratase EchA2 |
| M. smegmatis MC2 155 | MSMEG_1388 | - | 2e-50 | 37.70% (305) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_3498 | - | 2e-29 | 28.85% (260) | PUTATIVE enoyl-CoA hydratase/isomerase |
| M. ulcerans Agy99 | MUL_1414 | echA2 | 1e-162 | 90.43% (303) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_0780 | - | 1e-150 | 86.49% (296) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0464c|M.bovis_AF2122/97 -------MPTPDFQTLLYTTAGPVATITLNRPEQLNTIVPPMPDEIEAAI
Rv0456c|M.tuberculosis_H37Rv -------MPTPDFQTLLYTTAGPVATITLNRPEQLNTIVPPMPDEIEAAI
MAV_4694|M.avium_104 -------MPS-DFETLLYTTAGPVATITLNRPDQLNTIVPPMPDEIEAAV
MMAR_0778|M.marinum_M -------MPTPEFQTLLYSTAGPVATITLNRPEHLNTIVPPMPDEIEAAV
MUL_1414|M.ulcerans_Agy99 -------MPTPEFQTLLYSTAGPVATITLNRPEHLNTIVPPMPDEIEAAV
Mvan_0780|M.vanbaalenii_PYR-1 -------MPEPEFETLLYRADSAVATITLNRPEQLNTIVPPMPDEIEAAV
MAB_3859c|M.abscessus_ATCC_199 --------MVDTLKTMTYEVTDRVARITFNRPEQGNAIIADTPLELAALV
MSMEG_1388|M.smegmatis_MC2_155 MTHAIRPVDFDNLKTMTYEVTDRVARITFNRPEKGNAIVADTPLELSALV
Mflv_2433|M.gilvum_PYR-GCK -------------MYIDYEVSDRIATITLNRPEAANAQNPELLDELDAAW
TH_3498|M.thermoresistible__bu -------LETRTLEHVIYEKDGPIARIILNNPDRANAQTSEMVHSVNTAL
MLBr_02402|M.leprae_Br4923 ----------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAA
: :. * :*.*: *: . .:
Mb0464c|M.bovis_AF2122/97 GLAERDQDIKVIVLRGAGRAFSG-------GYDF----------GGGFQH
Rv0456c|M.tuberculosis_H37Rv GLAERDQDIKVIVLRGAGRAFSG-------GYDF----------GGGFQH
MAV_4694|M.avium_104 GRAERDPAIKVIVLRGAGRAFSG-------GYDF----------GGGFQH
MMAR_0778|M.marinum_M GLAERDQGIKVIVLRGAGRAFSG-------GYDF----------GGGFQH
MUL_1414|M.ulcerans_Agy99 GLAERDQGIKVIVLRGAGRAFSG-------GYDF----------GGGFQH
Mvan_0780|M.vanbaalenii_PYR-1 GLAERDPQVKVIVLRGAGRAFSG-------GYDF----------GGGFAH
MAB_3859c|M.abscessus_ATCC_199 ERADLDPQVHVILVSGRGDGFCG-------GFDLSAYAEAND--EGSVRY
MSMEG_1388|M.smegmatis_MC2_155 ERADLDPDVHVILVSGRGEGFCA-------GFDLSAYAEGSSSAGGGSPY
Mflv_2433|M.gilvum_PYR-GCK TRAAEDNDVSVIVLRANGKHFSA-------GHDLR---------------
TH_3498|M.thermoresistible__bu DDAQYDYDIKVVIIKANGKGFCSGHIPDGS--------------------
MLBr_02402|M.leprae_Br4923 KELDIDPDVGAILITGSPKVFAA---------------------------
* : .::: . *..
Mb0464c|M.bovis_AF2122/97 WGDAMM---------TDGRWDPGKDFAMVTARETGPTQKFMAIWR-ASKP
Rv0456c|M.tuberculosis_H37Rv WGDAMM---------TDGRWDPGKDFAMVTARETGPTQKFMAIWR-ASKP
MAV_4694|M.avium_104 WGEAMM---------TDGRWDPGKDFAMVSARETGPTQKFMAIWR-ASKP
MMAR_0778|M.marinum_M WGETMM---------TDGQWDPGKDFAMVTARETAPTQKFMAIWR-ASKP
MUL_1414|M.ulcerans_Agy99 WGETMM---------TDGQWDPGKDFAMVTARETAPTQKFMAIWR-ASKP
Mvan_0780|M.vanbaalenii_PYR-1 WGEAMN---------TDGRWDPGKDFAMVSARETGPTQKFMAIWR-ASKP
MAB_3859c|M.abscessus_ATCC_199 SGTALDGQVQLRNHLPNINWDPMLDYQMMSRFTRG----FSSLLH-ANKP
MSMEG_1388|M.smegmatis_MC2_155 EGTVLSGKTQALNHLPDEPWDPMVDYQMMSRFVRG----FASLMH-CDKP
Mflv_2433|M.gilvum_PYR-GCK ---------------GGGPVPDKITLEFIIQHEAKRYLEYTLRWRNVPKP
TH_3498|M.thermoresistible__bu ---------------YPEFKAEIEASGKVWRSAAQLFLWPVLKLWEFPKP
MLBr_02402|M.leprae_Br4923 ----------------GADIKEMASLTFTDAFDADFFSAWGKLAA-VRTP
.*
Mb0464c|M.bovis_AF2122/97 VIAQVHGWCVGGASDYALCADIVIASEDAVIGTPYSRM---WGAYLTGMW
Rv0456c|M.tuberculosis_H37Rv VIAQVHGWCVGGASDYALCADIVIASEDAVIGTPYSRM---WGAYLTGMW
MAV_4694|M.avium_104 VIAQVHGWCVGGASDYALCADLVIASEDAVIGTPYSRM---WGAYLTGMW
MMAR_0778|M.marinum_M VIAQIHGWCVGGASDYALCADIVIASDDAVIGTPYSRM---WGAYLSGMW
MUL_1414|M.ulcerans_Agy99 VIAQIHGWCVGGASDYALCADIVIASDDAVIGTPYSRM---WGAYLSGMW
Mvan_0780|M.vanbaalenii_PYR-1 VIAQVHGWCVGGASDYALCADIVIASDDAVIGTPYARM---WGAYLTGMW
MAB_3859c|M.abscessus_ATCC_199 TVVKVHGYCVAGGTDIALHADQLIVAADAKIGYPPTRV---WGVPATGLW
MSMEG_1388|M.smegmatis_MC2_155 TVVKIHGYCVAGGTDIALHADQVIAAADAKIGYPPMRV---WGVPAAGLW
Mflv_2433|M.gilvum_PYR-GCK SIAAVQGRCISGGLLLCWPCDLIVAADDAQFSDPVVLMG--IGGVEYHGH
TH_3498|M.thermoresistible__bu VIAQVHGYAIGGGTTWALIPEITVCSEDAWFQMPLVPGFGLPGSETMFEP
MLBr_02402|M.leprae_Br4923 MIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRL
:. : * .:.*. . : : : * : * *
Mb0464c|M.bovis_AF2122/97 LYRLSLAKVKWHSLTGRPLTGVQAAEAELINEAVPFERLEARVAEIATEL
Rv0456c|M.tuberculosis_H37Rv LYRLSLAKVKWHSLTGRPLTGVQAAEAELINEAVPFERLEARVAEIATEL
MAV_4694|M.avium_104 LYRLSLAKAKWHALTGRPLTGVQAAEIELINEAVPFERLEARVAEVAAEL
MMAR_0778|M.marinum_M LYRLSFAKAKWHSLTGRPLSGVQAAQIELINEAVPFERLEARVAEVAAEL
MUL_1414|M.ulcerans_Agy99 LYRLSFAKAKWHSLTGRPLSGVQAAQIELINEAVPFERLEARVAEVAAEL
Mvan_0780|M.vanbaalenii_PYR-1 IYRLSLAKVKWHSLTGEPLTGKEAAAVELINESVPFEQLEARVAEVAQKL
MAB_3859c|M.abscessus_ATCC_199 AHKLGDQRAKRLLLTGDCLSGRQAYDWGLAVEAPEPDELDERTERLVERI
MSMEG_1388|M.smegmatis_MC2_155 AHRLGDQRAKRLLFTGDCITGAQAAEWGLAVEAPDPADLDARTERLVERI
Mflv_2433|M.gilvum_PYR-GCK TWELGPRKAKEILFTGRAMTADEVAATGMVNKVVPRAELDAETRALAEQI
TH_3498|M.thermoresistible__bu WVFMNYKRAAEYLYTAQKITAQQALEFGLVNRVVPADELESTVEELAAKI
MLBr_02402|M.leprae_Br4923 TRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVATTI
:. :. *. : . :. : . * . :. :
Mb0464c|M.bovis_AF2122/97 ARIPLSQLQAQKLIVNQAYENMGLASTQLLGGILDGLMRNTPDALEFIRT
Rv0456c|M.tuberculosis_H37Rv ARIPLSQLQAQKLIVNQAYENMGLASTQLLGGILDGLMRNTPDALEFIRT
MAV_4694|M.avium_104 ARIPLSQLQAQKLIVNQAYENMGLASTQTLGGILDGLMRNTPDALEFIKI
MMAR_0778|M.marinum_M AQIPLSQLQAQKLIVNQAYENMGLASTQTLGGILDGLMRNTPDALDFIKT
MUL_1414|M.ulcerans_Agy99 AQIPLSQLQAQKLIVNQAYENMGLASTQTLGGILDGLMRNTPDALDFIKT
Mvan_0780|M.vanbaalenii_PYR-1 AGIPLSQLQAQKLIVNQAFENMGLASTQTLGGILDGLMRNTPDALGFIDT
MAB_3859c|M.abscessus_ATCC_199 AAMPLNQLMMIKLACNSVLINQGIANSAMIGTVFDGISRHTPEAHAFTAD
MSMEG_1388|M.smegmatis_MC2_155 AAMPVNQLIMAKLACNTALLNQGVATSQMVSTVFDGIARHTPEGHAFVAT
Mflv_2433|M.gilvum_PYR-GCK AKMPPFALRQAKRAVNQTLDVQGFY--AAIQSVFDVHQTGHGNALSVGGW
TH_3498|M.thermoresistible__bu AKAPLITLQATKAGILRAWETMGFRTHQQASNDLQAVVTGSREFQEYMAE
MLBr_02402|M.leprae_Br4923 SQMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFVTDDQSEGMAAF
: . * :. :
Mb0464c|M.bovis_AF2122/97 AQTQGVRAAVERRDGPFGDYSQAPPELRPDPTHVITPDGSM
Rv0456c|M.tuberculosis_H37Rv AQTQGVRAAVERRDGPFGDYSQAPPELRPDPTHVITPDGSM
MAV_4694|M.avium_104 AETRGVRAAVERRDGPFGDYSQAPPELRPDPSHVIVPDRDG
MMAR_0778|M.marinum_M AENQGVRAAVERRDGPFGDYSQAPPELRPNPNHVIVPDRDR
MUL_1414|M.ulcerans_Agy99 AENQGVRAAVERRDGPFGDYSQAPPELRPNPNHVIVPDRDR
Mvan_0780|M.vanbaalenii_PYR-1 AAADGVRAAVEQRDGPWGDYSQAPPHRRPDPSHVIEP----
MAB_3859c|M.abscessus_ATCC_199 AIANGYRQAVRHRDEPFGDYGRKPTEL--------------
MSMEG_1388|M.smegmatis_MC2_155 AREHGFREAVRRRDEPMGDHGRRASDV--------------
Mflv_2433|M.gilvum_PYR-GCK PVLVNLEEMKANIK---------------------------
TH_3498|M.thermoresistible__bu LMKRASKPADRV-----------------------------
MLBr_02402|M.leprae_Br4923 IEKRAPQFTHR------------------------------
.