For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AERPTPEEIILYDKDPETKIATITFNRPEFLNAPTSMARLRYADVLRAANADNDVKVVVIRGVGDNLGSG ADLPEFMEGNDNPAVRLAELRLEDDGVGEVTYPPRGTFRNGATISAWYANSQAGNRALQDFKKISIVEAK GYCYGWHFYQCADADLVISSDDALFGHPSFRYHGWGPRMWTWVQMMGLRKFQEMVFTGRPFTAAEMYDCN FLNKVVPRDRLEAEVDKYATACARNRPVDTVFQQKMFFEIFKQQQGEYMGSLLSAFFESMGNGVANDSDD DLDMFESIDSGLSAAVNDNDSRFPPEFRLSKKNRAKP*
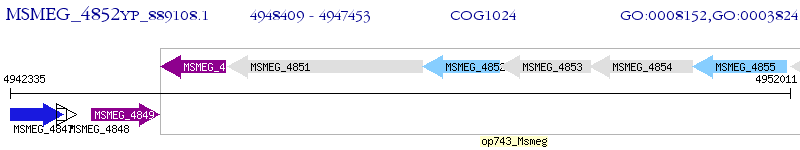
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4852 | - | - | 100% (318) | enoyl-CoA hydratase, putative |
| M. smegmatis MC2 155 | MSMEG_4846 | - | 5e-17 | 30.67% (225) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_1388 | - | 3e-14 | 24.63% (268) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0464c | echA2 | 4e-21 | 28.98% (314) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2422 | - | 0.0 | 96.20% (316) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv0456c | echA2 | 4e-21 | 28.98% (314) | enoyl-CoA hydratase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3859c | - | 2e-15 | 22.68% (291) | enoyl-CoA hydratase |
| M. marinum M | MMAR_4758 | echA4_1 | 1e-129 | 72.06% (315) | enoyl-CoA hydratase, EchA4_1 |
| M. avium 104 | MAV_2982 | - | 1e-102 | 56.41% (312) | enoyl-CoA hydratase/isomerase |
| M. thermoresistible (build 8) | TH_2025 | - | 1e-173 | 90.54% (317) | enoyl-CoA hydratase, putative |
| M. ulcerans Agy99 | MUL_1414 | echA2 | 1e-19 | 28.34% (314) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4230 | - | 0.0 | 95.89% (316) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2422|M.gilvum_PYR-GCK MAERPSPSAAERPTPEEIILYEKDPQTKIATITFNRPEFLNAPTSMARLR
Mvan_4230|M.vanbaalenii_PYR-1 MAERPQPPAAERPAPEEIILYDKDPKTKIATITFNRPEFLNAPTSMARLR
MSMEG_4852|M.smegmatis_MC2_155 --------MAERPTPEEIILYDKDPETKIATITFNRPEFLNAPTSMARLR
TH_2025|M.thermoresistible__bu --------MADRPAPEEIILYDKDPRTKIATITFNRPEFLNAPTSAARLR
MMAR_4758|M.marinum_M -------MPKSEPPVDKVIRYDKDPETRIATITFDRPGHLNAPTIAARLR
MAV_2982|M.avium_104 --MPEYDYEELKKEAEQYIKFEKDKKNRIAYITFDRPDAQNSATLGMRQN
Mb0464c|M.bovis_AF2122/97 ------------MPTPDFQTLLYTTAGPVATITLNRPEQLNTIVPPMPDE
Rv0456c|M.tuberculosis_H37Rv ------------MPTPDFQTLLYTTAGPVATITLNRPEQLNTIVPPMPDE
MUL_1414|M.ulcerans_Agy99 ------------MPTPEFQTLLYSTAGPVATITLNRPEHLNTIVPPMPDE
MAB_3859c|M.abscessus_ATCC_199 -------------MVDTLKTMTYEVTDRVARITFNRPEQGNAIIADTPLE
:* **::** *: .
Mflv_2422|M.gilvum_PYR-GCK YADVLRAANADNDVKVVIIRGVGDNLGSGADLPEFMEGNDNP--AVRLAE
Mvan_4230|M.vanbaalenii_PYR-1 YADVLRAANTDNDVKVVIIRGVGDNLGSGADLPEFMEGNDNP--AARLAE
MSMEG_4852|M.smegmatis_MC2_155 YADVLRAANADNDVKVVVIRGVGDNLGSGADLPEFMEGNDNP--AVRLAE
TH_2025|M.thermoresistible__bu YADLLRAATVDNDVKVVVIRGVGDNLGSGADLPEFMEGNDNP--ALRLAE
MMAR_4758|M.marinum_M YADLLHRASIDDEVKVLVIRGEGEDLGSGADLEEFMEVMNSPDQGPRLAE
MAV_2982|M.avium_104 YADLIHKCNVDDDVKVVVIRGEGEDFGSGGDLPEQRGMLENPG-MPLLHE
Mb0464c|M.bovis_AF2122/97 IEAAIGLAERDQDIKVIVLRGAGRAFSGGYDFG--------GGFQHWGDA
Rv0456c|M.tuberculosis_H37Rv IEAAIGLAERDQDIKVIVLRGAGRAFSGGYDFG--------GGFQHWGDA
MUL_1414|M.ulcerans_Agy99 IEAAVGLAERDQGIKVIVLRGAGRAFSGGYDFG--------GGFQHWGET
MAB_3859c|M.abscessus_ATCC_199 LAALVERADLDPQVHVILVSGRGDGFCGGFDLSAYAEANDEGSVRYSGTA
: . * ::*::: * * : .* *:
Mflv_2422|M.gilvum_PYR-GCK LRLEDDGVGEVTYPPKGTFRNGATISAWYANSQAGNRALQDFKKISIVEA
Mvan_4230|M.vanbaalenii_PYR-1 LRLEDDGVGEVTYPPKGTFRNGATISAWYANSQAGNRALQDFKKISIVEA
MSMEG_4852|M.smegmatis_MC2_155 LRLEDDGVGEVTYPPRGTFRNGATISAWYANSQAGNRALQDFKKISIVEA
TH_2025|M.thermoresistible__bu LRLEDDGVGEVTYPPKGTFRNGATISAWYANSQAGNRPLQELKKISIVEA
MMAR_4758|M.marinum_M FRIGPE---EVKYPPLGSFRHGASVSQWYANPNAGIRGLQDFKKISILEV
MAV_2982|M.avium_104 LAINDD---DVKYPPGGSYRYLSTVTDFYAKARAGNRPLQELRKISIIEA
Mb0464c|M.bovis_AF2122/97 M---------MTDGRWDPGKDFAMVTARETGPTQKFMAIWRASKPVIAQV
Rv0456c|M.tuberculosis_H37Rv M---------MTDGRWDPGKDFAMVTARETGPTQKFMAIWRASKPVIAQV
MUL_1414|M.ulcerans_Agy99 M---------MTDGQWDPGKDFAMVTARETAPTQKFMAIWRASKPVIAQI
MAB_3859c|M.abscessus_ATCC_199 LDGQVQLRNHLPNINWDPMLDYQMMSR----FTRGFSSLLHANKPTVVKV
: : .. :: : * : :
Mflv_2422|M.gilvum_PYR-GCK KGYCYGWHFYQCADADLVISSDDALFGHPSFRYHGWGPRMWTWVQMMGLR
Mvan_4230|M.vanbaalenii_PYR-1 KGYCYGWHFYQCADADLVISSDDALFGHPSFRYHGWGPRMWTWVQMMGLR
MSMEG_4852|M.smegmatis_MC2_155 KGYCYGWHFYQCADADLVISSDDALFGHPSFRYHGWGPRMWTWVQMMGLR
TH_2025|M.thermoresistible__bu KGYCYGWHFYQCADADLVISSDDALFGHPSFRYYGWGPRMWTWVQMMGLR
MMAR_4758|M.marinum_M KGYCYGWHFYQAADADLVISSDDALFGHPSFRYYGWGPRMWWWAQTMGIR
MAV_2982|M.avium_104 KGYCYGWHFYQAGDADLVIASDDALFGHPAFRYVGWGPRLWWWAETMGLR
Mb0464c|M.bovis_AF2122/97 HGWCVGGASDYALCADIVIASEDAVIGTPYSRMWG-AYLTGMWLYRLSLA
Rv0456c|M.tuberculosis_H37Rv HGWCVGGASDYALCADIVIASEDAVIGTPYSRMWG-AYLTGMWLYRLSLA
MUL_1414|M.ulcerans_Agy99 HGWCVGGASDYALCADIVIASDDAVIGTPYSRMWG-AYLSGMWLYRLSFA
MAB_3859c|M.abscessus_ATCC_199 HGYCVAGGTDIALHADQLIVAADAKIGYPPTRVWG-VPATGLWAHKLGDQ
:*:* . . ** :* : ** :* * * * * :.
Mflv_2422|M.gilvum_PYR-GCK KFQEMVFTGRPFTAAEMYDCNFLNKVVERDQLEAETQKYALACARNRPVD
Mvan_4230|M.vanbaalenii_PYR-1 KFQEMVFTGRPFTAAEMYDCNFLNKVVTRDQLEAETRKYALACARNRPVD
MSMEG_4852|M.smegmatis_MC2_155 KFQEMVFTGRPFTAAEMYDCNFLNKVVPRDRLEAEVDKYATACARNRPVD
TH_2025|M.thermoresistible__bu KFSEMVFTGRPFTAAEMYDCNFLNKVVPRDQLEAEVDKYARACARNRPVD
MMAR_4758|M.marinum_M KFQEMVFTGRAFTAAEMQDCNFLNSVVPREELEAEVAKYALACARNRPND
MAV_2982|M.avium_104 KFSEMLFTGRPFSAKEMYDCGFVNSVVPRDKLEAETEKYALACSKARPTD
Mb0464c|M.bovis_AF2122/97 KVKWHSLTGRPLTGVQAAEAELINEAVPFERLEARVAEIATELAR-IPLS
Rv0456c|M.tuberculosis_H37Rv KVKWHSLTGRPLTGVQAAEAELINEAVPFERLEARVAEIATELAR-IPLS
MUL_1414|M.ulcerans_Agy99 KAKWHSLTGRPLSGVQAAQIELINEAVPFERLEARVAEVAAELAQ-IPLS
MAB_3859c|M.abscessus_ATCC_199 RAKRLLLTGDCLSGRQAYDWGLAVEAPEPDELDERTERLVERIAA-MPLN
: . :** ::. : : : .. :.*: .. . . : * .
Mflv_2422|M.gilvum_PYR-GCK TVFQQKMFFEIFKQQQGEYMGSLLSAFFESMGNGVANDSDEDLDMFESID
Mvan_4230|M.vanbaalenii_PYR-1 TVFQQKMFFEIFKQQQGEYMGSLLSAFFESMGNGVANDSDDDLDMFESID
MSMEG_4852|M.smegmatis_MC2_155 TVFQQKMFFEIFKQQQGEYMGSLLSAFFESMGNGVANDSDDDLDMFESID
TH_2025|M.thermoresistible__bu TVFQQKIFFEVFKQHQGEYMGSLLSAFFESMGSGAVNDDTDDLDMHEAID
MMAR_4758|M.marinum_M TVFMQKVFFEIMKQFQGEYMGSLLSGFFESMGSGVRADT-DDLMLDDAIE
MAV_2982|M.avium_104 TVAVQKTFLELYKQHKGEYFGSLLTGLVEGMLPLIANDAEEVDLTAGTFE
Mb0464c|M.bovis_AF2122/97 QLQAQKLIVNQAYENMGLASTQLLGGILDGLMR---NTPDALEFIRTAQT
Rv0456c|M.tuberculosis_H37Rv QLQAQKLIVNQAYENMGLASTQLLGGILDGLMR---NTPDALEFIRTAQT
MUL_1414|M.ulcerans_Agy99 QLQAQKLIVNQAYENMGLASTQTLGGILDGLMR---NTPDALDFIKTAEN
MAB_3859c|M.abscessus_ATCC_199 QLMMIKLACNSVLINQGIANSAMIGTVFDGISR---HTPEAHAFTADAIA
: * : * : ..:.: :
Mflv_2422|M.gilvum_PYR-GCK SGLSAAVNDNDSKFPPEFRLSKRNRAKD-------------
Mvan_4230|M.vanbaalenii_PYR-1 SGLSAAVNDNDSKFPPEFRLSKRNRAKKD------------
MSMEG_4852|M.smegmatis_MC2_155 SGLSAAVNDNDSRFPPEFRLSKKNRAKP-------------
TH_2025|M.thermoresistible__bu SGLADAVNDNDMRFPPEFRLSKSNRAKE-------------
MMAR_4758|M.marinum_M QGLGDAVKDNDSRFPPEWRLSKKSRAKDGTGKPAKQKKKKG
MAV_2982|M.avium_104 KGLNNVVKDNDLNFPPEWRLSRSGRAKP-------------
Mb0464c|M.bovis_AF2122/97 QGVRAAVERRDGPFGDYSQAPPELRPDPTHVITPDGSM---
Rv0456c|M.tuberculosis_H37Rv QGVRAAVERRDGPFGDYSQAPPELRPDPTHVITPDGSM---
MUL_1414|M.ulcerans_Agy99 QGVRAAVERRDGPFGDYSQAPPELRPNPNHVIVPDRDR---
MAB_3859c|M.abscessus_ATCC_199 NGYRQAVRHRDEPFGDYGRKPTEL-----------------
.* .*. .* * : .