For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VWQDRLPQKYLEQGPRIVEENGMHLWHYDGQVFPTIGLNAVAGKPPEEWGMDPVRYEDMIPGCYDPVARV ADMDLDGVQSALCFPSFPGFGGGTFQRAEDKDLALLCVKAWNDFYIDEWCAVAPERYIPLAILPTWDIDA CVAEAERVAAKGARTISFPDSPVPLGLPSFHSDHWDGLWRVCSDAKMPVSLHFGSGSFVPGFSFSTIKPV PGQMAVPDAPFAVATALFSTNLMWSTVDLLFSGKLQQFPDLQISLAEGGIGWVPYILERTDYVWERHRYY QKIDFDTRPSDLFRKHFWGCFIDDEHGLTNRHSIGIDRIMLEIDFPHSDSNWPNSRKRAAEVLADVPDDE CRLIVEENARRMLNFPRVGAVDRQLAGV
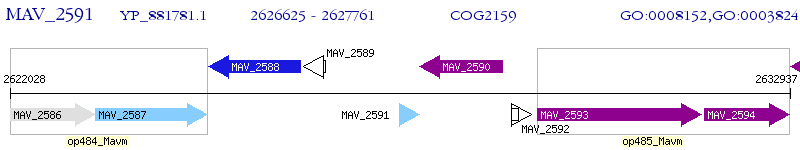
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2591 | - | - | 100% (378) | amidohydrolase 2 |
| M. avium 104 | MAV_1874 | - | e-146 | 89.73% (263) | amidohydrolase 2 |
| M. avium 104 | MAV_1837 | - | e-126 | 56.84% (373) | amidohydrolase 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2441 | - | 4e-84 | 39.48% (385) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4742 | - | 6e-88 | 40.36% (384) | metal-dependent amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_4837 | - | 2e-84 | 40.78% (385) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2339 | - | 3e-71 | 38.74% (364) | amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_3143 | - | 9e-74 | 38.59% (368) | amidohydrolase |
| M. vanbaalenii PYR-1 | Mvan_4211 | - | 2e-83 | 38.96% (385) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2441|M.gilvum_PYR-GCK ----------------MDKNDMILISVDDHIVEPPDMFKNHLSKKYLDEA
Mvan_4211|M.vanbaalenii_PYR-1 MSRAPARKGGNGRSAVMNRDDMILISVDDHIVEPPDMFKNHLPRKYLDEA
MMAR_4742|M.marinum_M ----------------MNKEDMILISVDDHTVEPPNMFKNHLSAKYLDDA
MSMEG_4837|M.smegmatis_MC2_155 ----------------MNKDDMILISVDDHIIEPPDMFKNHLPAKYRDEA
TH_2339|M.thermoresistible__bu ------------MSHDLRPEDLILVSIDDHVVEPPDMFLRHVPAKYRDEA
MUL_3143|M.ulcerans_Agy99 -----------MTDPPRARRRYTVISVDDHIVEPPDTFTGRLPQRFADRA
MAV_2591|M.avium_104 ------------------------------------MWQDRLPQKYLEQG
: ::. :: : .
Mflv_2441|M.gilvum_PYR-GCK PRLVHNEDGSDTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGC
Mvan_4211|M.vanbaalenii_PYR-1 PRLVHNPDGSDTWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGC
MMAR_4742|M.marinum_M PRLVHNPDGSDMWKFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGC
MSMEG_4837|M.smegmatis_MC2_155 PRLVHNPDGSDTWQFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIRKGC
TH_2339|M.thermoresistible__bu PIVVTDENGVDQWMYQGRPQGVSGLNAVVSWPPEEWGRNPARFAEMRPGV
MUL_3143|M.ulcerans_Agy99 PRVVETDDGGQVWVYDGQVLPNVGFNAVVGRPVSEYGFEPVRFDEMRRGA
MAV_2591|M.avium_104 PRIVE-ENGMHLWHYDGQVFPTIGLNAVAGKPPEEWGMDPVRYEDMIPGC
* :* :* . * : . .:***.. * .*:* :* :: *
Mflv_2441|M.gilvum_PYR-GCK WQVDERVKDMNAGGILGSMCFPSFP-GFAGR--LFATEDPEFSLALVQAY
Mvan_4211|M.vanbaalenii_PYR-1 WQVDERVKDMNAGGILGSMCFPSFP-GFAGR--LFATEDAEFSLALVQAY
MMAR_4742|M.marinum_M YNVDERIKDMNAGGILASICFPSFP-GFAGR--LFATEDPDFSVALVQAY
MSMEG_4837|M.smegmatis_MC2_155 YDPNERVKDMNAGGVLATMNFPSFP-GFAAR--LFATDDEEFSLALVQAY
TH_2339|M.thermoresistible__bu YDVHERVRDMNRNGILASMCFPTFT-GFSAR--HLNMHREHVTLIMVSAY
MUL_3143|M.ulcerans_Agy99 WDIHERIKDMDLNGVYASLNFPSFLPGFAGQRLQQVTKDPELAWAAVRAW
MAV_2591|M.avium_104 YDPVARVADMDLDGVQSALCFPSFP-GFGGG-TFQRAEDKDLALLCVKAW
:: *: **: .*: .:: **:* **.. . ..: * *:
Mflv_2441|M.gilvum_PYR-GCK NDWHVEEWCGAYPARFIPMTLPVIWDPEACAKEIRRNAARGVHSLTFSEN
Mvan_4211|M.vanbaalenii_PYR-1 NDWHVEEWCGAYPARFIPMTLPVIWDPEACAKEIRRNAARGVHSLTFSEN
MMAR_4742|M.marinum_M NDWHIDEWCGAYPARFVPMAIPVIWDAEACAAEVRRVAKKGVHALTFTEN
MSMEG_4837|M.smegmatis_MC2_155 NDWHIDEWCASNPGRFIPMAIPAIWNPGLAAAEVRRVAEKGVHSLTFTEN
TH_2339|M.thermoresistible__bu NDWHIDEWAGSYPGRFIPIAILPTWNPEAMCAEIRRVAAKGCRAVTMPEL
MUL_3143|M.ulcerans_Agy99 NDWHLEAWAGPYPERIIPCQLPWLLDPSAGAQLIHENAERGFRAVSFSEN
MAV_2591|M.avium_104 NDFYIDEWCAVAPERYIPLAILPTWDIDACVAEAERVAAKGARTISFPDS
**:::: *.. * * :* : : .. * :* :::::.:
Mflv_2441|M.gilvum_PYR-GCK PSAMGYPSFHDFDHWKPMWDALVDTDTVLNVHIGSSGRLA----------
Mvan_4211|M.vanbaalenii_PYR-1 PSAMGYPSFHDFEHWKPMWDALVDTDTVLNVHIGSSGRLA----------
MMAR_4742|M.marinum_M PAAMGYPSFHN-EYWNPLWKALCDTNTVMNIHIGSSGRLA----------
MSMEG_4837|M.smegmatis_MC2_155 PSTLGYPSFHDLEYWRPLWQALVDTETVMNVHIGSSGKLA----------
TH_2339|M.thermoresistible__bu PHLEGLPSYHDEDYWGPVFRTLSEENVVMCLHIGTGFGAI----------
MUL_3143|M.ulcerans_Agy99 PAMLGLPSIHS-GHWDPMMAACAETGTVVNLHIGSSGSSP----------
MAV_2591|M.avium_104 PVPLGLPSFHS-DHWDGLWRVCSDAKMPVSLHFGSGSFVPGFSFSTIKPV
* * ** *. :* : . : : :*:*:.
Mflv_2441|M.gilvum_PYR-GCK ---ITAPDAPMDVMITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEGGT
Mvan_4211|M.vanbaalenii_PYR-1 ---ITAPDAPMDVMITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEGGT
MMAR_4742|M.marinum_M ---ITAPDAPMDVMITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEGGT
MSMEG_4837|M.smegmatis_MC2_155 ---ITAPDAPMDVMITLQPMNIVQAAADLLWSAPIKEFPTLKIALSEGGT
TH_2339|M.thermoresistible__bu ---SMAPDAPIDNMIILATQVSAMCAQDLLWGPAMRNYPDLKFAFSEGGI
MUL_3143|M.ulcerans_Agy99 ---STTDDAPADVPGVLFFAYAISAAVDWLYSGLPSRFPDLKICLSEGRI
MAV_2591|M.avium_104 PGQMAVPDAPFAVATALFSTNLMWSTVDLLFSGKLQQFPDLQISLAEGGI
. *** * .: * *:. .:* *::.::**
Mflv_2441|M.gilvum_PYR-GCK GWIPYFLERVDRTYEMHSTWTG-QDFKGKLP-SEVFREHFLTCFIADPVG
Mvan_4211|M.vanbaalenii_PYR-1 GWIPYFLERVDRTYEMHSTWTG-QDFKGKLP-SEVFREHFLTCFIADPVG
MMAR_4742|M.marinum_M GWIPYFLERADRTFEMHSTWTH-QNFGGKIP-SEVFREHFLTCFISDKVG
MSMEG_4837|M.smegmatis_MC2_155 GWIPYFLDRVDRTFEMHSTWTH-QNFGGKLP-SEVFREHFMTCFISDPVG
TH_2339|M.thermoresistible__bu GWIPFYLDRCDRHYTN-QKWLR-RDFGDKLP-SDVFREHSLACYVTDKTS
MUL_3143|M.ulcerans_Agy99 GWVAGLLDRLDHMLSYHEMYGTWRAMGESLTSAEVFTRNFWFCAVEDKSS
MAV_2591|M.avium_104 GWVPYILERTDYVWERHRYYQK-IDFDTRPS--DLFRKHFWGCFIDDEHG
**:. *:* * : : . ::* .: * : * .
Mflv_2441|M.gilvum_PYR-GCK VATRHQIGVDNICWEADYPHSDSMWPGAPEQLDEVLKANNVPDDEINKMT
Mvan_4211|M.vanbaalenii_PYR-1 VATRHQIGVDNICWEADYPHSDSMWPGAPEQLDEVLKVNSVPDDEIDKMT
MMAR_4742|M.marinum_M VALRNMIGIDNIAWEADYPHSDSMWPGAPEELWDVLSVNDVPDGEINKMT
MSMEG_4837|M.smegmatis_MC2_155 VKNRHMIGIDNICWEMDYPHSDSMWPGAPEELSAVFDTYDVPDDEINKIT
TH_2339|M.thermoresistible__bu LKLRHEIGIDIIAWECDYPHSDCFWPDAPEQVHAELRAAGADDFDINKIT
MUL_3143|M.ulcerans_Agy99 FVQHYRIGTENIMVESDYPHCDSTWPHTQQTMHEQLDG--LPADVIRKIT
MAV_2591|M.avium_104 LTNRHSIGIDRIMLEIDFPHSDSNWPNSRKRAAEVLAD--VPDDECRLIV
. : ** : * * *:**.*. ** : : : :.
Mflv_2441|M.gilvum_PYR-GCK YENAMRWYHWDPFTHIAKEQATVGALRKAAEGHDVSIQALSNKE--KTGT
Mvan_4211|M.vanbaalenii_PYR-1 YQNAMRWYHWDPFTHIPKEQATVGALRKAAEGHDVSIQALSHKE--KTGA
MMAR_4742|M.marinum_M YENAMRWYSFDPFSHITREQATVGALRKAAEGHDVSVRALSHHKDARSGS
MSMEG_4837|M.smegmatis_MC2_155 HENAMRLYHFDPFAHVPKEQATVGALRKAAEGHDVSIRALSKKD--KTGA
TH_2339|M.thermoresistible__bu WENSCRFFSWDPFEHTPRAEATVGALRAKATDVDTTIRSR---------A
MUL_3143|M.ulcerans_Agy99 WENAARLYQHPVPVAIAQNPEAF---------------------------
MAV_2591|M.avium_104 EENARRMLNFPRVGAVDRQLAGV---------------------------
:*: * : .
Mflv_2441|M.gilvum_PYR-GCK TFSDFAASAKSISGNKD
Mvan_4211|M.vanbaalenii_PYR-1 TFADFAARAKEITGNTD
MMAR_4742|M.marinum_M SVAEFTANAKALTGNKD
MSMEG_4837|M.smegmatis_MC2_155 SFADFQANAKAVSGARD
TH_2339|M.thermoresistible__bu EWAQLYEQKQQFAQA--
MUL_3143|M.ulcerans_Agy99 -----------------
MAV_2591|M.avium_104 -----------------