For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRGKGYVVVGGTAGMGLAAARALAREGASVVVVGRDADRGKNAAEEVAAAGAASAHDLVFDVSQPGAAVA AVDEAVRLLGRLDGIAVTMGTAGMMPIDSDDDAWDNAFHDVLLATVRSVQAALPHLAVNGGAIVTTAAYS ARSPHEPRLPYASLKAAVATFTRGIARTHGKSGIRANSVAPGAVETDALHAIRGYVAESKGYPYAEALER ALVEDFGFDAALGRPGQPDEIGALIAFLLSDICAFVTGQTIYADGGAP
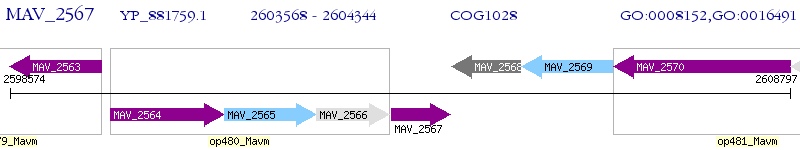
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2567 | - | - | 100% (258) | short-chain dehydrogenase/reductase SDR |
| M. avium 104 | MAV_1891 | - | e-118 | 82.56% (258) | short-chain dehydrogenase/reductase SDR |
| M. avium 104 | MAV_1587 | - | 3e-30 | 35.29% (255) | oxidoreductase, short chain dehydrogenase/reductase family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3589c | - | 2e-24 | 33.47% (251) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4353 | - | 6e-28 | 35.00% (260) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3559c | - | 3e-24 | 33.47% (251) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 2e-12 | 28.64% (199) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_0598 | - | 5e-26 | 33.33% (255) | short chain dehydrogenase |
| M. marinum M | MMAR_1568 | - | 2e-30 | 32.58% (264) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2228 | - | 2e-27 | 34.62% (260) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_3956 | - | 8e-49 | 43.85% (260) | PUTATIVE putative short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4844 | - | 1e-26 | 31.15% (260) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_5918 | - | 3e-26 | 32.69% (260) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3589c|M.bovis_AF2122/97 --------------------------------------------------
Rv3559c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0598|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5918|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2567|M.avium_104 --------------------------------------------------
TH_3956|M.thermoresistible__bu --------------------------------------------------
Mflv_4353|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2228|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1568|M.marinum_M --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mb3589c|M.bovis_AF2122/97 --------------------------------------------------
Rv3559c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0598|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5918|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2567|M.avium_104 --------------------------------------------------
TH_3956|M.thermoresistible__bu --------------------------------------------------
Mflv_4353|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2228|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1568|M.marinum_M --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mb3589c|M.bovis_AF2122/97 --------------------------------------------------
Rv3559c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0598|M.abscessus_ATCC_1997 --------------------------------------------------
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5918|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2567|M.avium_104 --------------------------------------------------
TH_3956|M.thermoresistible__bu --------------------------------------------------
Mflv_4353|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2228|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1568|M.marinum_M --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mb3589c|M.bovis_AF2122/97 ---------------------------------------------MNLSV
Rv3559c|M.tuberculosis_H37Rv ---------------------------------------------MNLSV
MAB_0598|M.abscessus_ATCC_1997 ---------------------------------------------MNLAE
MUL_4844|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5918|M.vanbaalenii_PYR-1 -------------------------------------------MNSAIVA
MAV_2567|M.avium_104 --------------------------------------------------
TH_3956|M.thermoresistible__bu --------------------------------------------------
Mflv_4353|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2228|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1568|M.marinum_M --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mb3589c|M.bovis_AF2122/97 APKEIAGHGLLDGKVVVVTAAAGTGIGSATARRALAEGADVVISDHHERR
Rv3559c|M.tuberculosis_H37Rv APKEIAGHGLLDGKVVVVTAAAGTGIGSATARRALAEGADVVISDHHERR
MAB_0598|M.abscessus_ATCC_1997 APTEIEGHGLLKGKAVVVTAAAGTGIGSSTARRALVEGADVVVSDYHERR
MUL_4844|M.ulcerans_Agy99 ------MTAELENKVAIITGACG-GIGLETSRVPACAGARVVLADLPETD
Mvan_5918|M.vanbaalenii_PYR-1 PEVPTAPIHDLTGKNAVITGGSK-GIGAATVARFVAGGARVVTSGRSQPD
MAV_2567|M.avium_104 ----------MRGKGYVVVGGTA-GMGLAAARALAREGASVVVVGRDADR
TH_3956|M.thermoresistible__bu ------MDLGLSGRNFVIVGGTT-GMGYAAAEQLARDGANVALLARDGDR
Mflv_4353|M.gilvum_PYR-GCK ------MDLGLRDATVVVVGGGR-GMGLAAAHCFADDGARVALVGRTRDV
MSMEG_2228|M.smegmatis_MC2_155 ------MDLGFGGAAAAVVGGGR-GMGFATAQCLADDGARVAVIGRSKDV
MMAR_1568|M.marinum_M ------MDLGFSGSKAVVTGGSK-GMGLAIAETLAAEGASVAVIARGKRA
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAAR-GIGAAIAEVVARDGARVVAIDVESAA
: . :... *:* ** *.
Mb3589c|M.bovis_AF2122/97 LGETAAELSALGLGRVEHVVCDVTS-TAQVDALIDSTTARMGRLDVLVNN
Rv3559c|M.tuberculosis_H37Rv LGETAAELSALGLGRVEHVVCDVTS-TAQVDALIDSTTARMGRLDVLVNN
MAB_0598|M.abscessus_ATCC_1997 LTETRDELAALGLGKVAAVVCDVTS-TEAVDALITESVAALGRIDVLVNN
MUL_4844|M.ulcerans_Agy99 LAGAAASVGRG----AVHHVVDLTN-EVSVRALIDFTIDTFGRLDIVDNN
Mvan_5918|M.vanbaalenii_PYR-1 SLPTG----------VEYVVADVAT-LDGVDLLARRAREILGDIDIIVNN
MAV_2567|M.avium_104 GKNAAEEVAAAGAASAHDLVFDVSQPGAAV-AAVDEAVRLLGRLDGIAVT
TH_3956|M.thermoresistible__bu ATERAAGLAEQHGVRAVGISVDAAATGDGVRHAIDEAAERLGPLRGLAVT
Mflv_4353|M.gilvum_PYR-GCK LDEAAAQLRGRGSPDAVGVVADCADEAAVQRAFGELAERWNGELNVLVNA
MSMEG_2228|M.smegmatis_MC2_155 LETAASDLARRGSPDAIAIVADTGDAEQVERAFAEIGERWDGELNVLINT
MMAR_1568|M.marinum_M LDSAVERLSQAGAPDAVAISADMADAASITSAFAAVSERWG-QLNCLVHT
MLBr_02565|M.leprae_Br4923 EALDETANRVGG----TTLSLDVTADDAVDKITEHLHDYQGGHADILVNN
* :
Mb3589c|M.bovis_AF2122/97 AGLGG--QTPVADMTDDEWDRVLDVSLTSVFRATRAALRYFRDAPHGGVI
Rv3559c|M.tuberculosis_H37Rv AGLGG--QTPVADMTDDEWDRVLDVSLTSVFRATRAALRYFRDAPHGGVI
MAB_0598|M.abscessus_ATCC_1997 AGLGG--ETPVADMTDDQWDRVLDVTLTSTFRATRAALRYFREAGHGGVI
MUL_4844|M.ulcerans_Agy99 AAHSDPADMLVSQMTVDVWDDTFTVNARGTMLMCKYAIPRLISAG-GGAI
Mvan_5918|M.vanbaalenii_PYR-1 AGATVPHLGGVLDIEDAEWVNDLNINFLAAVRLNAALLPALYARG-KGSI
MAV_2567|M.avium_104 MGTAGM-MP--IDSDDDAWDNAFHDVLLATVRSVQAALP-HLAVNGG-AI
TH_3956|M.thermoresistible__bu AGPMKQ-QGPFLEHADDSWDWYYQTILMATVRSCRAVIP-HLQRNGGGTI
Mflv_4353|M.gilvum_PYR-GCK VG-PTV-RGTFDELSDADWRQAVDEGAMGMVRCVRAALP-LLRAAEWARI
MSMEG_2228|M.smegmatis_MC2_155 VG-PGA-AGDFEELTDGQWQEAFDAGVMGMVRCVRAALP-LLRKAEWARI
MMAR_1568|M.marinum_M IG-PGD--GYFEELDDAAWDAAFTLGTMSGVRAIRAALP-LLRAADWARV
MLBr_02565|M.leprae_Br4923 AGITRD--KLLANMDDCRWDAVLEVNLLAPQRLTEGLVG-NGSISKGGRV
. : * . : :
Mb3589c|M.bovis_AF2122/97 VNNASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAAEYGVRINAVSPSI
Rv3559c|M.tuberculosis_H37Rv VNNASVLGWRAQHSQSHYAAAKAGVMALTRCSAIEAAEYGVRINAVSPSI
MAB_0598|M.abscessus_ATCC_1997 VNNASVLGWRAQYGQSHYAAAKAGVMALTRCSAIEAVEHGVRVNAVAPSI
MUL_4844|M.ulcerans_Agy99 VNISSATAHAAYDMSTAYSCTKAAIETLTRYVATQYGRHGVRCNAIAPGL
Mvan_5918|M.vanbaalenii_PYR-1 INVSSAVTLSPPSPMLHYATAKAALATYTTGLAAEAGPRGVRVNTLTPGT
MAV_2567|M.avium_104 VTTAAYSARSPHEPRLPYASLKAAVATFTRGIARTHGKSGIRANSVAPGA
TH_3956|M.thermoresistible__bu VTTAAYSVRAPKILIPPYNALKAAVLTLSKILAKTHGAEGIRVNTVCPGL
Mflv_4353|M.gilvum_PYR-GCK VNFSAHSTQRQSVILPAYTAAKAMVTSISKNLSLLLARDEILVNVVSPGS
MSMEG_2228|M.smegmatis_MC2_155 VNFSAHSTQRQSTRLPAYTAAKAAVNSVSKNLSLLLAKDEIMVNVVSPGS
MMAR_1568|M.marinum_M VTLSAHSIQRQNPRLVAYTASKAALSSVTKNLSKSLAKEGILVNCVCPGT
MLBr_02565|M.leprae_Br4923 IVLSSMAGIAGNRGQTNYATAKAGVIGLTQALAPGLAEKGITINAVAPGF
: :: * ** : : : : * : *.
Mb3589c|M.bovis_AF2122/97 ARHKFLDKTASAELLDRLVAGEAFG-----------------RAAEPWEV
Rv3559c|M.tuberculosis_H37Rv ARHKFLDKTASAELLDRLAAGEAFG-----------------RAAEPWEV
MAB_0598|M.abscessus_ATCC_1997 ARHKFLDKVTSSDLLDKLSSDEAFG-----------------RAAEPWEV
MUL_4844|M.ulcerans_Agy99 VRTPRLEVGLPQPIVDIFATHHLAG-----------------LIGEPHEI
Mvan_5918|M.vanbaalenii_PYR-1 VGSPGADAIRRAIIDGVGADRPEAGGIPIPLG----------RKGEASDI
MAV_2567|M.avium_104 VETDALHAIRGYVAESKGYPYAEALERALVED--FGFDAALGRPGQPDEI
TH_3956|M.thermoresistible__bu FDTEVNDFIRAQRAAEYNVAEEDAIYTHLSSNPAWNMRVALDRGGRPHEA
Mflv_4353|M.gilvum_PYR-GCK IA----SEALVGWADSVGVDGSDPYALMAAIDEHFGHPAHLPRAGLPTEI
MSMEG_2228|M.smegmatis_MC2_155 IS----SEALVGWAESVGVDGSDPYQLMAAIDEHFGHPAHLPRAGLPSEI
MMAR_1568|M.marinum_M IVTASFTEQLKDLLAADGLDATNPYDVMAWIDKYFHQPCDLGRAGLPEEV
MLBr_02565|M.leprae_Br4923 IETQMTAAIPLATREVGRRLNSLLQG------------------GLPVDV
. . :
Mb3589c|M.bovis_AF2122/97 AATIAFLASDYSSYLTGEVISVSCQHP---------------------
Rv3559c|M.tuberculosis_H37Rv AATIAFLASDYSSYLTGEVISVSCQHP---------------------
MAB_0598|M.abscessus_ATCC_1997 AATIAFLASDYSSYLTGEVISVSSQRA---------------------
MUL_4844|M.ulcerans_Agy99 AELVCFLASDRAAFITGQVIAADSGLLAHLPGLPQIRASVAELQSQPS
Mvan_5918|M.vanbaalenii_PYR-1 AEAIAFLASDRAAWITGSNLIVDGGQIQTR------------------
MAV_2567|M.avium_104 GALIAFLLSDICAFVTGQTIYADGGAP---------------------
TH_3956|M.thermoresistible__bu GELIAFLLSDRAAYTTGAAINIDGGTDF--------------------
Mflv_4353|M.gilvum_PYR-GCK GSVVAFLASRRNSYMTGADINVDGGSDFT-------------------
MSMEG_2228|M.smegmatis_MC2_155 GPVAAFLASRRNSYMTGANINVDGGSDFT-------------------
MMAR_1568|M.marinum_M ASITAYLVSRRNGYVTGATVNADGGSDFI-------------------
MLBr_02565|M.leprae_Br4923 AETIAYFAIPASNAVTGNVIRVCGQAMLGA------------------
. .:: ** :