For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPSANSSSLRDRRRAELLSQIQQTAHQLFAERGFEAVTTEDIAAAAGISISTYFRYAPTKEGLLVDPARE VAATIVGAYRTRPAHESAVEALIQLFVTHAKEVGAAGDFDTWRHAVATAPHLLRKSVLMSETDHSNLIQQ VAARMNVTAALDIRPALLVHTSLATVQFVLDGWLSSDSDESRPFHEQLEEALRITLAGFDRPSDATPGRR
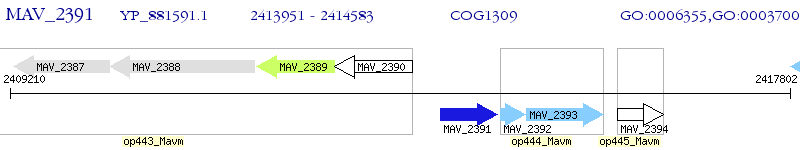
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2391 | - | - | 100% (210) | transcriptional regulator, TetR family protein |
| M. avium 104 | MAV_0350 | - | 3e-31 | 41.40% (186) | transcriptional regulator, TetR family protein |
| M. avium 104 | MAV_2512 | - | 8e-13 | 29.28% (181) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2534 | - | 1e-10 | 30.00% (180) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2551 | - | 6e-09 | 36.00% (100) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2506 | - | 1e-10 | 30.00% (180) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0248c | - | 8e-10 | 25.90% (166) | TetR family transcriptional regulator |
| M. marinum M | MMAR_3861 | - | 2e-69 | 67.84% (199) | putative regulatory protein |
| M. smegmatis MC2 155 | MSMEG_1420 | - | 2e-09 | 25.87% (201) | transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_3524 | - | 2e-07 | 25.28% (178) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_3793 | - | 1e-62 | 66.84% (187) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0989 | - | 1e-09 | 26.03% (146) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3861|M.marinum_M --MASEGDGMNPANIGSLRDRRRAELFSLIQQTAHRLFAERG-FDAVTTE
MUL_3793|M.ulcerans_Agy99 --MASEGDGMSPANIGSLRDRRRAELFSLIQQTAHRLFAERG-FDAVTTE
MAV_2391|M.avium_104 ---------MPSANSSSLRDRRRAELLSQIQQTAHQLFAERG-FEAVTTE
Mb2534|M.bovis_AF2122/97 MTASAPDGRPGQPEATNRRSQLKSDRRFQLLAAAERLFAERG-FLAVRLE
Rv2506|M.tuberculosis_H37Rv MTASAPDGRPGQPEATNRRSQLKSDRRFQLLAAAERLFAERG-FLAVRLE
Mflv_2551|M.gilvum_PYR-GCK ---MAP------TEISSPRSRAKSDRRDQLVAAAERLFAEHG-YLAVRLE
TH_3524|M.thermoresistible__bu -----------------VRGHRNRE---ALIAAAIDLFTVNG-YEQTTVE
MSMEG_1420|M.smegmatis_MC2_155 ------------MSEGSRAGRRRSTTQDHIAGVAIDLFAARG-FDAVSVD
MAB_0248c|M.abscessus_ATCC_199 -----------MRPASGLREQKKSETKLALSRAALELALDRGDLAAVTVD
Mvan_0989|M.vanbaalenii_PYR-1 -----------MTATPSLHEQRRRSTREALRRAALASFAARG-FANVTVT
: . : .* .* .
MMAR_3861|M.marinum_M DIAAAAGVSISTYFRHAPTKEGLLVDPVRQAITEIVSSYRSWPADES---
MUL_3793|M.ulcerans_Agy99 DIAAAAGVSISTYFRHAPTKEGLLVDPVRQAITEIVSSYRSWPADES---
MAV_2391|M.avium_104 DIAAAAGISISTYFRYAPTKEGLLVDPAREVAATIVGAYRTRPAHES---
Mb2534|M.bovis_AF2122/97 DIGAAAGVSGPAIYRHFPNKESLLVELLVGVSARLLAGARDVTTRSANLA
Rv2506|M.tuberculosis_H37Rv DIGAAAGVSGPAIYRHFPNKESLLVELLVGVSARLLAGARDVTTRSANLA
Mflv_2551|M.gilvum_PYR-GCK DIGSAAGISGPAVYRHFPNKEALLVELLVGISTRLLAGATDVVGADDDAA
TH_3524|M.thermoresistible__bu QIAEAAGVAPRTFFRHFAAKEDILFHGYADRLAEATRRFR--AARSGSLW
MSMEG_1420|M.smegmatis_MC2_155 DVAAAAGISRRTLFRYYASKSAIPWGDFDSHLQHLQNLLRTLSSEVSLGD
MAB_0248c|M.abscessus_ATCC_199 DIADAVRVSARTFRNYFTSKEDAVLFSLRGIEDNAVALLRQRPQHEHVVD
Mvan_0989|M.vanbaalenii_PYR-1 ELARAAGVTERTFFRHFPTKEAVLFQDYETQVEWLAAALARRPGSESVFD
::. *. :: : .: . *.
MMAR_3861|M.marinum_M -AVEALIALFVSYARDAGDL-KLDTWRRAIATAPYLLSKSALVSEDDQHR
MUL_3793|M.ulcerans_Agy99 -AVEASIALFVSYARDAGDL-KLDTLRRAIATAPYLLSKSALVSEDDQHR
MAV_2391|M.avium_104 -AVEALIQLFVTHAKEVGAAGDFDTWRHAVATAPHLLRKSVLMSETDHSN
Mb2534|M.bovis_AF2122/97 AALDGLIEFHLDFALGEADL--IRIQDRDLAHLPAVAERQVRKAQRQYVE
Rv2506|M.tuberculosis_H37Rv AALDGLIEFHLDFALGEADL--IRIQDRDLAHLPAVAERQVRKAQRQYVE
Mflv_2551|M.gilvum_PYR-GCK STLENLIDFHLDFALDESDL--IRIQDRDLGNLPPAAKRQVRRKQRQYVE
TH_3524|M.thermoresistible__bu KALAEASEAVATAIAENPGI--FLVRARMYGELPALRATMLRINDDWIDQ
MSMEG_1420|M.smegmatis_MC2_155 ALREALLTFNTYADHEMAEH---RQRMRVILETEELQAYSMTMYAGWRTV
MAB_0248c|M.abscessus_ATCC_199 SLEAVMLDLATSEAFSTTVA-----VTRLSAQNPGLAAHDAARSEDVGTV
Mvan_0989|M.vanbaalenii_PYR-1 AVRAAIAEFPYDLEVVRQAA----TARSELISAERIAGHLRVVQSSFADV
MMAR_3861|M.marinum_M FIEHVASRMGVDARTDIRPALLVHTSLATVKFVFDRWLSTDPPSSPIFHV
MUL_3793|M.ulcerans_Agy99 FIEHVASRMGVDARTDIRPALLVHTSLATVKFVFDRWLSTDPPSSPIFHV
MAV_2391|M.avium_104 LIQQVAARMNVTAALDIRPALLVHTSLATVQFVLDGWLSSDSDESRPFHE
Mb2534|M.bovis_AF2122/97 VWVGVLRELNP-GLAEADARLMAHAVFGLLNSTPHSMKAADSKPARTVRA
Rv2506|M.tuberculosis_H37Rv VWVGVLRELNP-GLAEADARLMAHAVFGLLNSTPHSMKAADSKPARTVRA
Mflv_2551|M.gilvum_PYR-GCK IWVDVLRRCTP-GLSEADARLMAHGTFGLLNSTAHSMKPGTTKTAEAG-S
TH_3524|M.thermoresistible__bu MTTEVARWLGSDAAVDVRPRLAATVINGANRTAIDLWVSADGKGDLVELM
MSMEG_1420|M.smegmatis_MC2_155 IAEYVADRLGM-STTDLMPQTVAWLMLGVALSAYENWLADESVSLRGAIA
MAB_0248c|M.abscessus_ATCC_199 VLTEVSRRAGLDPDVDLYPRLVCNAAYAVLATVIQLAVSGSKLSKDPG--
Mvan_0989|M.vanbaalenii_PYR-1 LTGHIRTRYAGTADIDLVADVAGAALAAALVVAVEHWGRNGCVDDLGEIT
. : : . . . .
MMAR_3861|M.marinum_M QMDQ-----------------------------------------ALRIA
MUL_3793|M.ulcerans_Agy99 QWTRRCGSRSPDSGNAIRHLPSSRETEPNFVVRHDIWVRPSGSGSNPRAS
MAV_2391|M.avium_104 QLEE-----------------------------------------ALRIT
Mb2534|M.bovis_AF2122/97 RAVLR------------------------------------------AMT
Rv2506|M.tuberculosis_H37Rv RAVLR------------------------------------------AMT
Mflv_2551|M.gilvum_PYR-GCK RTVLR------------------------------------------EMT
TH_3524|M.thermoresistible__bu ADAVR----------------------------------------LVRPT
MSMEG_1420|M.smegmatis_MC2_155 AAFEV-----------------------------------------ARPG
MAB_0248c|M.abscessus_ATCC_199 -----------------------------------------------ALV
Mvan_0989|M.vanbaalenii_PYR-1 SASMA-------------------------------------------VL
MMAR_3861|M.marinum_M LAGFR----------------
MUL_3793|M.ulcerans_Agy99 ACSFERMRPCIHHRRHHCTR-
MAV_2391|M.avium_104 LAGFDRPSDATPGRR------
Mb2534|M.bovis_AF2122/97 VAALSAADRCL----------
Rv2506|M.tuberculosis_H37Rv VAALSAADRCL----------
Mflv_2551|M.gilvum_PYR-GCK VAALTSAERRRG---------
TH_3524|M.thermoresistible__bu LDRIERESRTVANPVRRRDVV
MSMEG_1420|M.smegmatis_MC2_155 LDALQPAH-------------
MAB_0248c|M.abscessus_ATCC_199 AQGFQRLRDGLA---------
Mvan_0989|M.vanbaalenii_PYR-1 RSGFARLT-------------
: