For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MEYGMRPELAEFRAEVRAFVAEHAPAIPPRAGVRSAENDAEFKALQEWTGRLFEAGYVGADWPVEFGGRG DFTPEHSIIVGEELARAQAPSAQSGSVLASHALIHYGTDEQRRRHLPEIRGGRELWCQLFSEPGAGSDLA SLRTRAVRHGDTYTIDGQKVWTTDGHWARYGYLLARTDPDAPKHKGISAFILDMSAPGVSVRPLRELTGT SDFNEVFFDGVEVPADAMIGAPGQGWAIANATLAHERTNVGGIVVKLRLAIDALIDLARRVTIDGGPAID SDRVRDRIGQFSAEVEALSALTYANLTRWLRGRERMHDAAMGKLMFSEINLEMAAFAVELAGEAGVLVEG DDNVLDGGRWQDEWLYARAYTIAGGSSEIMRNLIAERGLGLPRD
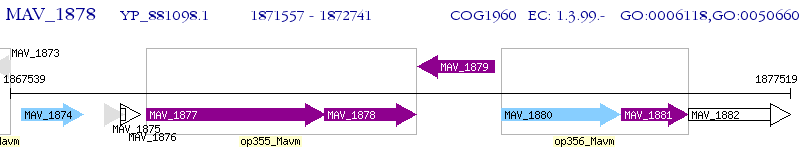
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1878 | - | - | 100% (394) | acyl-CoA dehydrogenase family protein |
| M. avium 104 | MAV_2594 | - | 0.0 | 84.26% (394) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_1268 | - | 8e-74 | 37.44% (398) | acyl-CoA dehydrogenase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3590c | fadE30 | 9e-69 | 39.59% (394) | acyl-CoA dehydrogenase FADE30 |
| M. gilvum PYR-GCK | Mflv_1794 | - | 1e-67 | 40.97% (393) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3560c | fadE30 | 9e-69 | 39.59% (394) | acyl-CoA dehydrogenase FADE30 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 3e-33 | 31.96% (341) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0597 | - | 2e-66 | 38.62% (391) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_2862 | fadE17 | 1e-68 | 39.60% (404) | acyl-CoA dehydrogenase FadE17 |
| M. smegmatis MC2 155 | MSMEG_5619 | - | 9e-68 | 40.82% (392) | acyl-CoA dehydrogenase domain-containing protein |
| M. thermoresistible (build 8) | TH_2543 | - | 7e-98 | 47.41% (386) | PUTATIVE putative acyl-CoA dehydrogenase family protein |
| M. ulcerans Agy99 | MUL_4125 | fadE30 | 2e-64 | 37.97% (395) | acyl-CoA dehydrogenase FadE30 |
| M. vanbaalenii PYR-1 | Mvan_4956 | - | 4e-68 | 41.31% (397) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1878|M.avium_104 -----------MEYGMRP-----------ELAEFRAEVRAFVAEHAPAIP
TH_2543|M.thermoresistible__bu -----------VGLVTRPTAQ--------HLDEFRARVRTFIAENAPPFQ
Mb3590c|M.bovis_AF2122/97 --------------MQDV-------------EEFRAQVRGWLADNLAGEF
Rv3560c|M.tuberculosis_H37Rv --------------MQDV-------------EEFRAQVRGWLADNLAGEF
MUL_4125|M.ulcerans_Agy99 --------------MQDV-------------EEFRAEVRGWLAANLVGEF
MAB_0597|M.abscessus_ATCC_1997 MVESQVSPAPAGEPMSEMSD-----------DQVRTEIREWLAENLVGEF
MMAR_2862|M.marinum_M -----------MDVAYPPEA-----------EEFRERVRAFLDEHLPCGW
Mflv_1794|M.gilvum_PYR-GCK --------------MSELET-------------FRAEVRDWCREHVPADW
Mvan_4956|M.vanbaalenii_PYR-1 -------------MTAESEDQRDNPTDSQSQAQFRQQVRAWCRDHIPADW
MSMEG_5619|M.smegmatis_MC2_155 --------------MTHPEN----------LDDFRATVRDWCAAHVPKDW
MLBr_00737|M.leprae_Br4923 ----------MVGWSGNP-----------LFDLFKLPEEHNELRATIRAL
.: .
MAV_1878|M.avium_104 PRAG--VRSAENDAEFKALQEWTGRLFEAGYVGADWPVEFGGRGDFTPEH
TH_2543|M.thermoresistible__bu AREG--HRAPVDATQEKQLRTWFAALFEAKLIGADWPVEHGGRADHHPLY
Mb3590c|M.bovis_AF2122/97 AALKGLGGPGREHEAFEERRAWNQRLAAAGLTCLGWPEEHGGR-GLSTAH
Rv3560c|M.tuberculosis_H37Rv AALKGLGGPGREHEAFEERRAWNQRLAAAGLTCLGWPEEHGGR-GLSTAH
MUL_4125|M.ulcerans_Agy99 ATLKGLGGPGREHEAFEERRAWNQHLAAAGLTCLGWPVEHGGR-GLSTAH
MAB_0597|M.abscessus_ATCC_1997 AELKGLGGPGSEHEAFEERLAWNRHLAAAGWTCLGWPVEHGGR-GATVSQ
MMAR_2862|M.marinum_M SGPGALAVQDR--KAFAAR--WRQLLVAAGLIAVSWPKEYGGG-GYSPVE
Mflv_1794|M.gilvum_PYR-GCK RAAQ-SGVGEEEFVRFQKA--WFAELHSAGYAVPHWPAQWGG--GLPVDK
Mvan_4956|M.vanbaalenii_PYR-1 RDTQ-TGVGDEEFVRFQKA--WFAELHSAGYAVPHWPAEWGG--GLPVDR
MSMEG_5619|M.smegmatis_MC2_155 RAAQ-TGVSDDEFAAFQKA--WFAELHSAGYAVPHWPAEWGG--GMSVPQ
MLBr_00737|M.leprae_Br4923 AEKEIAPHAADVDQRARFPEEALAALNASGFNAIHVPEEYGGQ-GADSVA
* : * : ** .
MAV_1878|M.avium_104 SIIVGEELARAQAPSAQSG----SVLASHALIHYGTDEQRRRHLPEIRGG
TH_2543|M.thermoresistible__bu DRIVSEEILRARAPRPVDQ----VNLAAHVLLHFGSDEQKRTYLPAIRRS
Mb3590c|M.bovis_AF2122/97 RVAFYEEYARADAPDKVNH--FGEELLGPTLIAFGTPQQQRRFLPRIRDV
Rv3560c|M.tuberculosis_H37Rv RVAFYEEYARADAPDKVNH--FGEELLGPTLIAFGTPQQQRRFLPRIRDV
MUL_4125|M.ulcerans_Agy99 RVAFYEEYARADAPDKVNH--FGEELLGPTLIAFGTPEQQQRFLPRILDV
MAB_0597|M.abscessus_ATCC_1997 RVIFYEEYARAEAPHKVNH--LGEELLGPTLIAYGTPEQQQRFLPAIRDV
MMAR_2862|M.marinum_M QVVLAEEFARAGAPEREENDLFGVDLLGNTLIALGTEAQKRHFLPRILTG
Mflv_1794|M.gilvum_PYR-GCK QVVLYQELAAHDAPRLVLA-FVGIHHAASTLLVAGTEEQRRRHLPAILDG
Mvan_4956|M.vanbaalenii_PYR-1 QVVLYQELAAHDAPRLVLA-FVGIHHAASTLLVAGTDEQRRRHLPAILDG
MSMEG_5619|M.smegmatis_MC2_155 QIVLYSELAAHDAPRLVLA-FVGIHHAASTLLAAGTDAQRNRHLPAILDG
MLBr_00737|M.leprae_Br4923 ACIVIEEVARVDASASLIP--AVNKLGTMGLILRGSEELKKQVLPSLAAE
. .* *. *: *: :. ** :
MAV_1878|M.avium_104 RELWCQLFSEPGAGSDLASLRTRAVRHGD-TYTIDGQKVWTTDGHWARYG
TH_2543|M.thermoresistible__bu EHVWCQLLSEPDAGSDIAAVRSRAVRLPDGTWRVDGQKTWITDGHWADMG
Mb3590c|M.bovis_AF2122/97 TELWCQGYSEPGAGSDLASVATTAELDGD-QWVINGQKVWTSLAHLSQWC
Rv3560c|M.tuberculosis_H37Rv TELWCQGYSEPGAGSDLASVATTAELDGD-QWVINGQKVWTSLAHLSQWC
MUL_4125|M.ulcerans_Agy99 TELWCQGYSEPGAGSDLANVATTAELDGD-QWVINGQKVWTSLAHLSQWC
MAB_0597|M.abscessus_ATCC_1997 TELWCQGYSEPGAGSDLANVSTSAVLDGG-DWVINGQKVWTSLALQSQWC
MMAR_2862|M.marinum_M DDRWCQGFSEPEAGSDLASVRTRAVLDGD-QWVINGQKIWTSAGSTANWI
Mflv_1794|M.gilvum_PYR-GCK -EIWVQGFSEPEAGSDLAALRTSARRVGD-EFIVNGQKLWASGGMHADWC
Mvan_4956|M.vanbaalenii_PYR-1 -EIWVQGFSEPEAGSDLAALRTTARRAGD-EFVVNGQKLWASGGMHADWC
MSMEG_5619|M.smegmatis_MC2_155 -EIWVQGFSEPEAGSDLASLRTTARAEGD-TFVVNGQKLWASGAMYADWC
MLBr_00737|M.leprae_Br4923 GAMASYALSEREAGSDAASMRTRAKADGD-DWILNGFKCWITNGGKSTWY
** **** * : : * . : ::* * * : . :
MAV_1878|M.avium_104 YLLARTDPDAPKH------KGISAFILDMSAPGVSVRPLRELTGT--SDF
TH_2543|M.thermoresistible__bu LALLRTDPAAVRH------HGLSVFVVPLSAPGVEVRPIRTIGDA--IEV
Mb3590c|M.bovis_AF2122/97 FVLARTEKGSQRH------AGLSYLLVPLDQPGVQIRPIVQITGT--AEF
Rv3560c|M.tuberculosis_H37Rv FVLARTEKGSQRH------AGLSYLLVPLDQPGVQIRPIVQITGT--AEF
MUL_4125|M.ulcerans_Agy99 FVVARTEKGSKRH------AGLSYLLVPLDQPSVQIRPIVQITGT--AEF
MAB_0597|M.abscessus_ATCC_1997 FVVARTEPGSSRH------KGLSYLLVPLDQPGVEIRPIIQLTGT--SEF
MMAR_2862|M.marinum_M FVLARTDPVAPKH------QGLSLLLVPMDQPAVVVRPIVNAAGH--SLF
Mflv_1794|M.gilvum_PYR-GCK LLLARTDPDAPKR------KGISYFLLDMTSPGVDVRPIRNAIGD--SHF
Mvan_4956|M.vanbaalenii_PYR-1 LLLARTDPDAPKRPGARKSAGISYFLMDMTTPGVDVRPIRNAIGD--SHF
MSMEG_5619|M.smegmatis_MC2_155 LLLARTDPNAPKR------HGISYFMMDMKTPGVDVRPIRNAVGD--SHF
MLBr_00737|M.leprae_Br4923 TVMAVTDPDKGAN-------GISAFIVHKDDEGFSIGPKEKKLGIKGSPT
: *: . *:* ::: .. : * .
MAV_1878|M.avium_104 NEVFFDGVEVPADAMIGAPGQGWAIANATLAHER-TNVGGIVVKLRLAID
TH_2543|M.thermoresistible__bu NEVFLTDVRLPAENLIGAEGQGWSIIMAGLDFER-FGIGGNVILLELLID
Mb3590c|M.bovis_AF2122/97 NEVFFDDARTDADLVVGAPGDGWRVAMATLTFERGVSTLGQQIVYARELS
Rv3560c|M.tuberculosis_H37Rv NEVFFDDARTDADLVVGAPGDGWRVAMATLTFERGVSTLGQQIVYARELS
MUL_4125|M.ulcerans_Agy99 NEVFFDDARTDADLVVGAPGDGWRVAMGTLTFERGVSTLGQQIVYARELS
MAB_0597|M.abscessus_ATCC_1997 NEVFFDNARTSADLVVGEPGDGWRVAMGTLTFERGVSTLGNQITYARELS
MMAR_2862|M.marinum_M NEVFFADARTGVANVVGEVGSGWTAAMSLLGFERGAQATTGAIDFARDLS
Mflv_1794|M.gilvum_PYR-GCK CEIFLDDVRIPATNLIGAENAGWQVAQETLGAERGMTMLELAERLG--NA
Mvan_4956|M.vanbaalenii_PYR-1 CEIFLDDVRIPAANLIGAENNGWQVAQATLGAERGLTMLELAERLG--NA
MSMEG_5619|M.smegmatis_MC2_155 CEIFLNDVVIPSANLVGPVNKGWQVAQATLGAERGMTMLELAERLG--NA
MLBr_00737|M.leprae_Br4923 TELYFDKCRIPGDRIIGEPGTGFKTALATLDHTRPTIGAQAVGIAQGALD
*::: ::* . *: * *
MAV_1878|M.avium_104 ALIDLARRVTIDGGPAIDSDRVRDRIGQFSAEVEALSALTYANLTRWLRG
TH_2543|M.thermoresistible__bu DLLTLARHLQVDGRPAIEVADIRQRIAELAVEVEVAKAFIDDHVEQLING
Mb3590c|M.bovis_AF2122/97 NLVELARRTAAADDPLI-----RERLTRAWTGLRAMR--SYALATMEGPA
Rv3560c|M.tuberculosis_H37Rv NLVELARRTAAADDPLI-----RERLTRAWTGLRAMR--SYALATMEGPA
MUL_4125|M.ulcerans_Agy99 DLAALARRTGAADDPFI-----REHLTRAWTGLRAMR--SYALATMD---
MAB_0597|M.abscessus_ATCC_1997 RLAELAKSNGAADDPAI-----RERLAQAYVGMRTMR--AYALATMD---
MMAR_2862|M.marinum_M RLCDLARARGLDSDPRI-----RDRLAWCYSRTQIMRYRGYRSLTWALNG
Mflv_1794|M.gilvum_PYR-GCK GFRWLVES-APVDDPIV-----ADRLAGFETELAALRGMCRRMVEH---G
Mvan_4956|M.vanbaalenii_PYR-1 GFRWLVQS-APVDDPIV-----ADRLAGFETEITGLRGLCRQIVER---G
MSMEG_5619|M.smegmatis_MC2_155 GFRWLVDACSPVDDPVV-----ADRLAQFEIELTGLRGLCRDLVERSETG
MLBr_00737|M.leprae_Br4923 AAIVYTKDRKQFGESISTFQSIQFMLADMAMKVEAARLIVYAAAARAERG
. . . :
MAV_1878|M.avium_104 RERMHDA-AMGKLMFSEINLEMAAFAVELAGEAGVLVEGDD---------
TH_2543|M.thermoresistible__bu REQPGDS-SIAKLSFAETYHTVAAYGADLASWGTSFQPADS---------
Mb3590c|M.bovis_AF2122/97 VEQPGQD-NVSKLLWANWHRNLGELAMDVIGKPGMTMP------------
Rv3560c|M.tuberculosis_H37Rv VEQPGQD-NVSKLLWANWHRNLGELAMDVIGKPGMTMP------------
MUL_4125|M.ulcerans_Agy99 VEQPGQD-NVSKLLWANWHRSLGELAMDVIGKPGLTLA------------
MAB_0597|M.abscessus_ATCC_1997 EERPGQD-NVSKLLWANWHRDLGELAMEIIGTSTLLTN------------
MMAR_2862|M.marinum_M LLPGAEA-AISKVIWSEYFQRYTELAVEILGLDALCPSGPGNGEARVVPQ
Mflv_1794|M.gilvum_PYR-GCK PSGPADP-SVVKLYYSELLQRLTDFGAEIGGLPAHTVLTKP---------
Mvan_4956|M.vanbaalenii_PYR-1 ATGPADA-SVVKLYYSELLQRLTDFAVEIGGPEAHTVLAKP---------
MSMEG_5619|M.smegmatis_MC2_155 EAGPADA-SIVKLYYSELLQRMTDFGAQIAGLPAHTVLTKP---------
MLBr_00737|M.leprae_Br4923 EPDLGFISAASKCFASDIAMEVTTDAVQLFGGAGYTSDFP----------
* :: . :: .
MAV_1878|M.avium_104 -NVLDG-GRWQDEWLYARAYTIAGGSSEIMRNLIAERGLGLPRD------
TH_2543|M.thermoresistible__bu -PVAQARQRLRECWLWSRAYTISGGSSEMMRNILAKRRLKLPTT------
Mb3590c|M.bovis_AF2122/97 ---DGEFDEWQRLYLFTRADTIYGGSNEIQRNIIAERVLGLPREAKG---
Rv3560c|M.tuberculosis_H37Rv ---DGEFDEWQRLYLFTRADTIYGGSNEIQRNIIAERVLGLPREAKG---
MUL_4125|M.ulcerans_Agy99 ---DGEFDEWQRLFLFTRADTIYGGSNEIQRNIIAERVLGLPREAKG---
MAB_0597|M.abscessus_ATCC_1997 ---AGEMDEWQRLFLFSRADTIYGGSNEIQRNIISERVLGLPREAR----
MMAR_2862|M.marinum_M AGTANSPACWMDELLYARAATIYAGSSQIQRNVIGEQLLGLPKEPRREAV
Mflv_1794|M.gilvum_PYR-GCK MSSGWESGSWVLDFVGSWEWTIPGGASEIQRTIIGERGLGLPREPSAL--
Mvan_4956|M.vanbaalenii_PYR-1 MSSGWESGSWVLDFVGSWEWTIPGGASEIQRTIIAERGLGLPREPGAA--
MSMEG_5619|M.smegmatis_MC2_155 ASSGWESGAWMLDFIGSWEWTIPGGASEIQRTIIGERGLGLPREPSAV--
MLBr_00737|M.leprae_Br4923 ---------VERFMRDAKITQIYEGTNQIQRVVMSRALLR----------
: * *:.:: * ::.. *
MAV_1878|M.avium_104 -
TH_2543|M.thermoresistible__bu -
Mb3590c|M.bovis_AF2122/97 -
Rv3560c|M.tuberculosis_H37Rv -
MUL_4125|M.ulcerans_Agy99 -
MAB_0597|M.abscessus_ATCC_1997 -
MMAR_2862|M.marinum_M H
Mflv_1794|M.gilvum_PYR-GCK -
Mvan_4956|M.vanbaalenii_PYR-1 -
MSMEG_5619|M.smegmatis_MC2_155 -
MLBr_00737|M.leprae_Br4923 -