For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPLQDHHQIVSVDDHLVEHPRVWQDRLPEKWREQGPRIVELDGKHLWSYDGQVFPTIGLNAVAGKPPEEW GLDPVRYEDMIPGCYDPVARVADMDLDGVQAALCFPSFPGFGGGTFVRAEVNRPGFDGGSCYWISTSGWS VRFVA
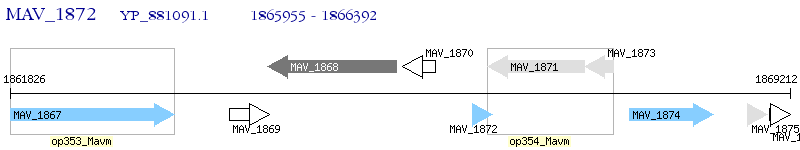
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1872 | - | - | 100% (145) | amidohydrolase 2 |
| M. avium 104 | MAV_2591 | - | 2e-52 | 89.90% (99) | amidohydrolase 2 |
| M. avium 104 | MAV_1837 | - | 7e-45 | 60.32% (126) | amidohydrolase 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2441 | - | 2e-25 | 44.04% (109) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2618 | - | 1e-28 | 50.46% (109) | amidohydrolase |
| M. smegmatis MC2 155 | MSMEG_4837 | - | 9e-27 | 45.61% (114) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2339 | - | 1e-23 | 45.19% (104) | amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_3143 | - | 8e-29 | 50.46% (109) | amidohydrolase |
| M. vanbaalenii PYR-1 | Mvan_4211 | - | 2e-26 | 43.86% (114) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2441|M.gilvum_PYR-GCK -------------------------------MDKNDMILISVDDHIVEPP
Mvan_4211|M.vanbaalenii_PYR-1 ---------------MSRAPARKGGNGRSAVMNRDDMILISVDDHIVEPP
MSMEG_4837|M.smegmatis_MC2_155 -------------------------------MNKDDMILISVDDHIIEPP
TH_2339|M.thermoresistible__bu ---------------------------MSHDLRPEDLILVSIDDHVVEPP
MMAR_2618|M.marinum_M MKSIDELASDLSFTTAKTGADRSVTFLPDPPRARRRYTVISVDDHIVEPP
MUL_3143|M.ulcerans_Agy99 --------------------------MTDPPRARRRYTVISVDDHIVEPP
MAV_1872|M.avium_104 ------------------------------MPLQDHHQIVSVDDHLVEHP
::*:***::* *
Mflv_2441|M.gilvum_PYR-GCK DMFKNHLSKKYLDEAPRLVHNEDGSDTWQFRDVVIPNVALNAVAGRPKEE
Mvan_4211|M.vanbaalenii_PYR-1 DMFKNHLPRKYLDEAPRLVHNPDGSDTWQFRDVVIPNVALNAVAGRPKEE
MSMEG_4837|M.smegmatis_MC2_155 DMFKNHLPAKYRDEAPRLVHNPDGSDTWQFRDTVIPNVALNAVAGRPKEE
TH_2339|M.thermoresistible__bu DMFLRHVPAKYRDEAPIVVTDENGVDQWMYQGRPQGVSGLNAVVSWPPEE
MMAR_2618|M.marinum_M DTFTGRLPQRFADRAPRVVETDDGGQVWVYDGQVLPNVGFNAVVGRPVSE
MUL_3143|M.ulcerans_Agy99 DTFTGRLPQRFADRAPRVVETDDGGQVWVYDGQVLPNVGFNAVVGRPVSE
MAV_1872|M.avium_104 RVWQDRLPEKWREQGPRIVEL-DGKHLWSYDGQVFPTIGLNAVAGKPPEE
: ::. :: :..* :* :* . * : . .:***.. * .*
Mflv_2441|M.gilvum_PYR-GCK YGLEPQGLDEIRPGCWQVDERVKDMNAGGILGSMCFPSFP-GFAGR--LF
Mvan_4211|M.vanbaalenii_PYR-1 YGLEPQGLDEIRPGCWQVDERVKDMNAGGILGSMCFPSFP-GFAGR--LF
MSMEG_4837|M.smegmatis_MC2_155 YGLEPQGLDEIRKGCYDPNERVKDMNAGGVLATMNFPSFP-GFAAR--LF
TH_2339|M.thermoresistible__bu WGRNPARFAEMRPGVYDVHERVRDMNRNGILASMCFPTFT-GFSAR--HL
MMAR_2618|M.marinum_M YGFEPVRFDEMRRGAWDIHERIKDMDLNGVYASLNFPSFLPGFAGQRLQQ
MUL_3143|M.ulcerans_Agy99 YGFEPVRFDEMRRGAWDIHERIKDMDLNGVYASLNFPSFLPGFAGQRLQQ
MAV_1872|M.avium_104 WGLDPVRYEDMIPGCYDPVARVADMDLDGVQAALCFPSFP-GFGGG----
:* :* :: * :: *: **: .*: .:: **:* **..
Mflv_2441|M.gilvum_PYR-GCK ATEDPEFSLALVQAYNDWHVEEWCGAYPARFIPMTLPVIWDPEACAKEIR
Mvan_4211|M.vanbaalenii_PYR-1 ATEDAEFSLALVQAYNDWHVEEWCGAYPARFIPMTLPVIWDPEACAKEIR
MSMEG_4837|M.smegmatis_MC2_155 ATDDEEFSLALVQAYNDWHIDEWCASNPGRFIPMAIPAIWNPGLAAAEVR
TH_2339|M.thermoresistible__bu NMHREHVTLIMVSAYNDWHIDEWAGSYPGRFIPIAILPTWNPEAMCAEIR
MMAR_2618|M.marinum_M VTKDPELAWAAVRAWNDWHLEAWAGPYPERIIPCQLPWLLDPSAGAQLIH
MUL_3143|M.ulcerans_Agy99 VTKDPELAWAAVRAWNDWHLEAWAGPYPERIIPCQLPWLLDPSAGAQLIH
MAV_1872|M.avium_104 ----------------------------------------------TFVR
::
Mflv_2441|M.gilvum_PYR-GCK RNAARGVHSLTFSENPSAMGYPSFHDFDHWKPMWDALVDTDTVLNVHIGS
Mvan_4211|M.vanbaalenii_PYR-1 RNAARGVHSLTFSENPSAMGYPSFHDFEHWKPMWDALVDTDTVLNVHIGS
MSMEG_4837|M.smegmatis_MC2_155 RVAEKGVHSLTFTENPSTLGYPSFHDLEYWRPLWQALVDTETVMNVHIGS
TH_2339|M.thermoresistible__bu RVAAKGCRAVTMPELPHLEGLPSYHDEDYWGPVFRTLSEENVVMCLHIGT
MMAR_2618|M.marinum_M ENAERGFRAVSFSENPAMLGLPSIHS-GHWDPMMAACAETGTVVNLHIGS
MUL_3143|M.ulcerans_Agy99 ENAERGFRAVSFSENPAMLGLPSIHS-GHWDPMMAACAETGTVVNLHIGS
MAV_1872|M.avium_104 AEVNR----------------PGFDGGS----------------CYWIST
. : *. .. *.:
Mflv_2441|M.gilvum_PYR-GCK SGRLAITAPDAPMDVMITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEG
Mvan_4211|M.vanbaalenii_PYR-1 SGRLAITAPDAPMDVMITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEG
MSMEG_4837|M.smegmatis_MC2_155 SGKLAITAPDAPMDVMITLQPMNIVQAAADLLWSAPIKEFPTLKIALSEG
TH_2339|M.thermoresistible__bu GFGAISMAPDAPIDNMIILATQVSAMCAQDLLWGPAMRNYPDLKFAFSEG
MMAR_2618|M.marinum_M SGSSPSTTDDAPADVPGVLFFAYAISAAVDWLYSGLPSRFPDLKICLSEG
MUL_3143|M.ulcerans_Agy99 SGSSPSTTDDAPADVPGVLFFAYAISAAVDWLYSGLPSRFPDLKICLSEG
MAV_1872|M.avium_104 S-------------------------------------------------
.
Mflv_2441|M.gilvum_PYR-GCK GTGWIPYFLERVDRTYEMHSTWTG-QDFKGKLP-SEVFREHFLTCFIADP
Mvan_4211|M.vanbaalenii_PYR-1 GTGWIPYFLERVDRTYEMHSTWTG-QDFKGKLP-SEVFREHFLTCFIADP
MSMEG_4837|M.smegmatis_MC2_155 GTGWIPYFLDRVDRTFEMHSTWTH-QNFGGKLP-SEVFREHFMTCFISDP
TH_2339|M.thermoresistible__bu GIGWIPFYLDRCDRHYTN-QKWLR-RDFGDKLP-SDVFREHSLACYVTDK
MMAR_2618|M.marinum_M GIGWVAGLLDRLDHMLSYHEMYGTWRAMGESLTPAEVFTRNFWFCAVEDK
MUL_3143|M.ulcerans_Agy99 RIGWVAGLLDRLDHMLSYHEMYGTWRAMGESLTSAEVFTRNFWFCAVEDK
MAV_1872|M.avium_104 --GWSVRFVA----------------------------------------
** :
Mflv_2441|M.gilvum_PYR-GCK VGVATRHQIGVDNICWEADYPHSDSMWPGAPEQLDEVLKANNVPDDEINK
Mvan_4211|M.vanbaalenii_PYR-1 VGVATRHQIGVDNICWEADYPHSDSMWPGAPEQLDEVLKVNSVPDDEIDK
MSMEG_4837|M.smegmatis_MC2_155 VGVKNRHMIGIDNICWEMDYPHSDSMWPGAPEELSAVFDTYDVPDDEINK
TH_2339|M.thermoresistible__bu TSLKLRHEIGIDIIAWECDYPHSDCFWPDAPEQVHAELRAAGADDFDINK
MMAR_2618|M.marinum_M SSFVQHHRIGTENIMVESDYPHCDSTWPHTQQTMHEQLDG--LPADVIRK
MUL_3143|M.ulcerans_Agy99 SSFVQHYRIGTENIMVESDYPHCDSTWPHTQQTMHEQLDG--LPADVIRK
MAV_1872|M.avium_104 --------------------------------------------------
Mflv_2441|M.gilvum_PYR-GCK MTYENAMRWYHWDPFTHIAKEQATVGALRKAAEGHDVSIQALSNKEKTGT
Mvan_4211|M.vanbaalenii_PYR-1 MTYQNAMRWYHWDPFTHIPKEQATVGALRKAAEGHDVSIQALSHKEKTGA
MSMEG_4837|M.smegmatis_MC2_155 ITHENAMRLYHFDPFAHVPKEQATVGALRKAAEGHDVSIRALSKKDKTGA
TH_2339|M.thermoresistible__bu ITWENSCRFFSWDPFEHTPRAEATVGALRAKATDVDTTIRSR-------A
MMAR_2618|M.marinum_M ITWENAARLYQHPVPVAIAQNPEAF-------------------------
MUL_3143|M.ulcerans_Agy99 ITWENAARLYQHPVPVAIAQNPEAF-------------------------
MAV_1872|M.avium_104 --------------------------------------------------
Mflv_2441|M.gilvum_PYR-GCK TFSDFAASAKSISGNKD
Mvan_4211|M.vanbaalenii_PYR-1 TFADFAARAKEITGNTD
MSMEG_4837|M.smegmatis_MC2_155 SFADFQANAKAVSGARD
TH_2339|M.thermoresistible__bu EWAQLYEQKQQFAQA--
MMAR_2618|M.marinum_M -----------------
MUL_3143|M.ulcerans_Agy99 -----------------
MAV_1872|M.avium_104 -----------------