For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRRTIVITGASDGIGAAAARRLSRAGDRIVVVGRSQTKTAAVAAELGADHFVVDYADLSQVRALADKMR AQYPRIDVLLNNAGGVASRIELTADGYERTYQVNYLAPFLLTTQLLDVLLESRATVVNTTSSSHKLILRA TVDDLENTANRRPAVAYAYSKLAIVLFTKELHRRYHARGLSVAAVHPGNVNSNIGIASGSRFLVFMQRYT PAALFISSPDQGADPLVRLASSPPDSEWTSGAYYAKRKIGKTTRLADDPRLAAELWERTAARLG
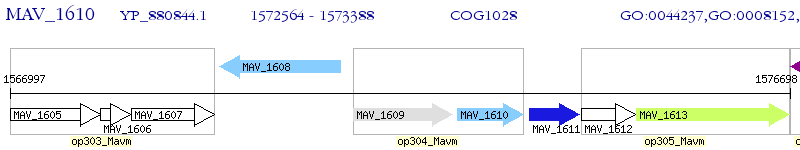
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1610 | - | - | 100% (274) | NAD dependent epimerase/dehydratase family protein |
| M. avium 104 | MAV_1829 | - | 1e-63 | 51.13% (266) | retinol dehydrogenase 14 |
| M. avium 104 | MAV_4710 | - | 8e-30 | 36.11% (288) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0447c | - | 2e-28 | 34.88% (258) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0143 | - | 2e-30 | 35.27% (258) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0439c | - | 2e-28 | 34.88% (258) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 2e-25 | 34.39% (253) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3438 | - | 2e-32 | 34.75% (282) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_3998 | - | 1e-101 | 67.15% (274) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6557 | - | 3e-28 | 36.55% (238) | dehydrogenase |
| M. thermoresistible (build 8) | TH_4179 | - | 2e-27 | 35.61% (264) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3862 | - | 7e-99 | 65.69% (274) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0762 | - | 3e-33 | 36.61% (254) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0447c|M.bovis_AF2122/97 MTANDNKTRKWS---AADVPDQSGRVVVVTGANTGIGYHTAAVFADRGAH
Rv0439c|M.tuberculosis_H37Rv MTANDNKTRKWS---AADVPDQSGRVVVVTGANTGIGYHTAAVFADRGAH
MLBr_00315|M.leprae_Br4923 -------MAKWT---TADIPDQTGRVAVITGANTGLGYQTALALAEHGAH
TH_4179|M.thermoresistible__bu ------MTGKWT---ARDVPDQTGRTAVITGANTGLGFETAKVLAEKGAH
Mflv_0143|M.gilvum_PYR-GCK -------MTDWTNWNVADIPDQTGRTAVITGANTGLGFETAKALAEKGAR
Mvan_0762|M.vanbaalenii_PYR-1 -------MTDWT---TADIPDQTGRTAVITGANTGLGFETAKALAAGGAR
MSMEG_6557|M.smegmatis_MC2_155 ------MTSTWS---PDRLGDLGDKRIIVTGATNGVGLGTTRALVRAGAH
MMAR_3998|M.marinum_M ---------------------MTGKTIVITGASDGIGAAAARRLTQNGEK
MUL_3862|M.ulcerans_Agy99 ---------------------MTGKTIVITGASDGIGAGAARRLTQNGEK
MAV_1610|M.avium_104 ---------------------MTRRTIVITGASDGIGAAAARRLSRAGDR
MAB_3438|M.abscessus_ATCC_1997 -------------MSTYPDRPVAGSRIVITGATNGIGKEIARALVRRGAL
::***. *:* : : *
Mb0447c|M.bovis_AF2122/97 VVLAVRNLEKGNAARARIMAARPG-AHVTLQQLDLCSLDSVRAAADALRT
Rv0439c|M.tuberculosis_H37Rv VVLAVRNLEKGNAARARIMAARPG-AHVTLQQLDLCSLDSVRAAADALRT
MLBr_00315|M.leprae_Br4923 VVLAVRNLDKGKDAAARITATSAQ-NNVALQELDLASLESVRAAAKQLRS
TH_4179|M.thermoresistible__bu VVLAVRDPDKGRRAADRITAAAPH-ADVTVRQLDLTSLDNIRRAADDLRA
Mflv_0143|M.gilvum_PYR-GCK VVIAVRNTDKGAQAAARIRG------DVDVQELDLTSLSSIRTAAEALKA
Mvan_0762|M.vanbaalenii_PYR-1 VVLAVRDTEKGAQAAAKMAG------DVDVQQLDLTSLASIRSAADALKS
MSMEG_6557|M.smegmatis_MC2_155 VILAVRNTDLGARRAAEVGG------TTTVVEVDLADMSSVRAFPGRLTA
MMAR_3998|M.marinum_M VVVVGRSESKTTAVAAQLRAD--------YFVADFADLSQVRALADKIRS
MUL_3862|M.ulcerans_Agy99 VVVVGRSESKTTAVAAQLRAD--------YFVADFADLSQVRALADKIRS
MAV_1610|M.avium_104 IVVVGRSQTKTAAVAAELGAD--------HFVVDYADLSQVRALADKMRA
MAB_3438|M.abscessus_ATCC_1997 LTLLARDPAKASATVRELAAENGAITAPEYIQADLADLDSVRAAAHEIAT
: : *. .: . * .: .:* . : :
Mb0447c|M.bovis_AF2122/97 A-YPRIDVLINNAGVMWTPKQVTKDGFELQFGTNHLGHFALTGLVLDHML
Rv0439c|M.tuberculosis_H37Rv A-YPRIDVLINNAGVMWTPKQVTKDGFELQFGTNHLGHFALTGLVLDHML
MLBr_00315|M.leprae_Br4923 D-YDHIDLLINNAGVMWTPKSTTKDGFELQFGTNHLGHFAFTGLLLDRLL
TH_4179|M.thermoresistible__bu G-YPRIDLLINNAGVMYPPRQTTRDGFELQFGTNHLGHFALTGQLLDNIL
Mflv_0143|M.gilvum_PYR-GCK R-FDKIDLLINNAGVMTTPKGTTADGFELQFGTNHLGHFALTGLLFDNIL
Mvan_0762|M.vanbaalenii_PYR-1 R-FDHIDLLINNAGVMTTPKGTTADGFELQFGTNHLGHFAFTGLLLDKLL
MSMEG_6557|M.smegmatis_MC2_155 AGIDDVDILINNAGALTQRRTETVDGFETTLGTNLLGPFALTNLLLGRVR
MMAR_3998|M.marinum_M E-YPRIDVLANNAGAMCHEIQFTADGYEKTYQVNYLSPFLLTTLLLD-VL
MUL_3862|M.ulcerans_Agy99 E-YPRIDVLANNAGAMCHEFQLTADAYEKTYQVNYLSSFLLTTLRLD-VL
MAV_1610|M.avium_104 Q-YPRIDVLLNNAGGVASRIELTADGYERTYQVNYLAPFLLTTQLLD-VL
MAB_3438|M.abscessus_ATCC_1997 T-HPRIDTLVNNAGIHSLSGRTSADGFDLMTATNHLGPFLLTNLLLEPIV
:* * **** : *.:: .* *. * :* : :
Mb0447c|M.bovis_AF2122/97 PVPGSRVVTVSSQGHRIHAAIHFDDLQWERR---YNRVAAYGQAKLANLL
Rv0439c|M.tuberculosis_H37Rv PVPGSRVVTVSSQGHRIHAAIHFDDLQWERR---YNRVAAYGQAKLANLL
MLBr_00315|M.leprae_Br4923 PIVGSRVITVSSLSHRLFADIHFNDLQWECN---YNRVAAYGQSKLANLL
TH_4179|M.thermoresistible__bu PVDGSRVVTVASIAHRNMADIHFDDLQWERG---YHRVAAYGQSKLANLM
Mflv_0143|M.gilvum_PYR-GCK DIPGSRIVTVSSNGHKMGGAIHWDDLQWERS---YNRMGAYTQSKLANLL
Mvan_0762|M.vanbaalenii_PYR-1 DVDGARIVTVSSNGHKMGGAIHWDDLQWERS---YSRMGAYSQSKLANLL
MSMEG_6557|M.smegmatis_MC2_155 ----SQVIIVGSDAHR-MATLRLDDMHLRRNR--WTPLGAYGRAKLAVML
MMAR_3998|M.marinum_M VQSRATVVNTTSSSQRLLPKVTLAELESTAQR---RPSVAYALTKLAIVL
MUL_3862|M.ulcerans_Agy99 VQSRATVVNTTSSSQRLLPKVTLAELENTAQR---RPSVAYALTKLAIVL
MAV_1610|M.avium_104 LESRATVVNTTSSSHKLILRATVDDLENTANR---RPAVAYAYSKLAIVL
MAB_3438|M.abscessus_ATCC_1997 AAERARIVITASEAHRSWPRIDLERFAEARRYNAIRSEIRYGQSKLMNIL
: :: . * .:: : * :** ::
Mb0447c|M.bovis_AF2122/97 FTYELQRRLGEAGKSTIAVAAHPGGSNTELT----RNLPRLIRPVATVLG
Rv0439c|M.tuberculosis_H37Rv FTYELQRRLGEAGKSTIAVAAHPGGSNTELT----RNLPRLIRPVATVLG
MLBr_00315|M.leprae_Br4923 FTYELQRRLATR-QTTIAVAAHPGGSRTELT----RTLPALIAPIFSVAE
TH_4179|M.thermoresistible__bu FAYELQRRLSAKNAPTISVAAHPGVSNTELT----RYIPGARLPGVSLLA
Mflv_0143|M.gilvum_PYR-GCK FTYELQRRLAPR-GKTIAVAAHPGTSTTELA----RNLPRPVERAFLAAA
Mvan_0762|M.vanbaalenii_PYR-1 FTYELQRRLAPR-GKTIAVAAHPGTSSTELG----RNLPAALQPALNRLA
MSMEG_6557|M.smegmatis_MC2_155 WGLELDRRQRVARSPVVTHLTHPGWVASNLPNVSQTRLMSVIQRGVTTVA
MMAR_3998|M.marinum_M FTRELHRRYHAK--GLSAATFHPGYVDSNFGEASGSRALAFMKYHVPLVA
MUL_3862|M.ulcerans_Agy99 FTRELHRRYHAK--GLSAATFHPGYVDSNFGEASGSRALAFMKYHVPLVT
MAV_1610|M.avium_104 FTKELHRRYHAR--GLSVAAVHPGNVNSNIGIASGSRFLVFMQRYTP-AA
MAB_3438|M.abscessus_ATCC_1997 FTQELSRRLDGT--GVTANCFCPGMVST--GLVRDSRVLTAVAGLAS-RT
: ** ** ** :
Mb0447c|M.bovis_AF2122/97 P-LLFQSPEMGALPTLR-AATDPTTQGGQYYGPDGFGEQRGHPKVVQSSA
Rv0439c|M.tuberculosis_H37Rv P-LLFQSPEMGALPTLR-AATDPTTQGGQYYGPDGFGEQRGHPKVVQSSA
MLBr_00315|M.leprae_Br4923 L-FLTQDAATGALPTLR-AATDAAVLGGQYFGPDGFAEIRGHPKVVASNG
TH_4179|M.thermoresistible__bu G-LLTNSPAVGALATLR-AATDPEVKGGQYYGPDGFQEIRGHPVLVGSSA
Mflv_0143|M.gilvum_PYR-GCK PVLFAQTADRGALPTLR-AAADPGVLGGQYYGPDGLGQQRGAPVVVASSA
Mvan_0762|M.vanbaalenii_PYR-1 P-LLAQSPAAGALPTLR-AATDPSVLGGQYYGPDGIGQQRGNPVVVASSN
MSMEG_6557|M.smegmatis_MC2_155 D-RFAASIDEGAAPTLY-CISEP-IPPGSYVGVEGRLGLRGAPVLIGRSA
MMAR_3998|M.marinum_M RLTSTAEQGAEQLVWLASSAAGTDWTSGEYYSRHKIAK---------ANR
MUL_3862|M.ulcerans_Agy99 RFTSTAEQGAEQLVWLASSAAGTDWTSGEYYSRHKIAR---------ANR
MAV_1610|M.avium_104 LFISSPDQGADPLVRLASSPPDSEWTSGAYYAKRKIGK---------TTR
MAB_3438|M.abscessus_ATCC_1997 PFVRRPDQGARMGLRLVLDEDLATTTGQFFTSTPGLGQL-------PAVR
* . : .
Mb0447c|M.bovis_AF2122/97 QSHDKDLQRRLWTVSEELTGVSFGV------------------------
Rv0439c|M.tuberculosis_H37Rv QSHDKDLQRRLWTVSEELTGVSFGV------------------------
MLBr_00315|M.leprae_Br4923 KSHDVDRQLRLWAVSEELTGVVYPVG-----------------------
TH_4179|M.thermoresistible__bu KSRDEDIQRRLWTVSEELTGVTFPV------------------------
Mflv_0143|M.gilvum_PYR-GCK QSYDVDQQRRLWEISEELTKVTFPDL-----------------------
Mvan_0762|M.vanbaalenii_PYR-1 QSYDMALQRRLWTVSEELTGVTFPVPVLS--------------------
MSMEG_6557|M.smegmatis_MC2_155 VACDYETAAAVVDFAERETGTAIPY------------------------
MMAR_3998|M.marinum_M AAYDPRLASELWDRTLAKLR-----------------------------
MUL_3862|M.ulcerans_Agy99 AAYDPRLASELWDRTLAKLRREARPATSGIHQIVKSIMPLTMSPLSRSV
MAV_1610|M.avium_104 LADDPRLAAELWERTAARLG-----------------------------
MAB_3438|M.abscessus_ATCC_1997 IRSDQKAQEELWRRSGELVSL----------------------------
* : :