For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGKTIVITGASDGIGAAAARRLTQNGEKVVVVGRSESKTTAVAAQLRADYFVADFADLSQVRALADKIRS EYPRIDVLANNAGAMCHEIQFTADGYEKTYQVNYLSPFLLTTLLLDVLVQSRATVVNTTSSSQRLLPKVT LAELESTAQRRPSVAYALTKLAIVLFTRELHRRYHAKGLSAATFHPGYVDSNFGEASGSRALAFMKYHVP LVARLTSTAEQGAEQLVWLASSAAGTDWTSGEYYSRHKIAKANRAAYDPRLASELWDRTLAKLR*
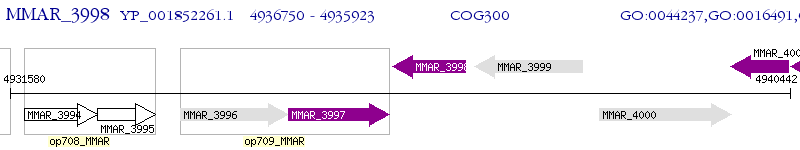
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3998 | - | - | 100% (275) | short chain dehydrogenase |
| M. marinum M | MMAR_0757 | - | 4e-27 | 33.68% (285) | dehydrogenase/reductase |
| M. marinum M | MMAR_1680 | - | 1e-24 | 33.71% (264) | dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0069 | - | 2e-26 | 32.27% (282) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0143 | - | 1e-25 | 36.13% (191) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0068 | - | 2e-26 | 32.27% (282) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 8e-25 | 33.45% (284) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3438 | - | 2e-32 | 35.42% (288) | putative short-chain dehydrogenase/reductase |
| M. avium 104 | MAV_1610 | - | 1e-101 | 67.15% (274) | NAD dependent epimerase/dehydratase family protein |
| M. smegmatis MC2 155 | MSMEG_0876 | - | 1e-25 | 30.88% (285) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4179 | - | 4e-24 | 33.59% (256) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3862 | - | 1e-147 | 96.36% (275) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0762 | - | 8e-27 | 30.96% (281) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0069|M.bovis_AF2122/97 MT-KWT---AADIPDQTGRTAVITGANTGLGFETAAALAAHGAHVVLAVR
Rv0068|M.tuberculosis_H37Rv MT-KWT---AADIPDQTGRTAVITGANTGLGFETAAALAAHGAHVVLAVR
MSMEG_0876|M.smegmatis_MC2_155 MT-KWT---TARIPDQTGRVAIVTGANTGLGLETAKALAAKGAHVVLAVR
MLBr_00315|M.leprae_Br4923 MA-KWT---TADIPDQTGRVAVITGANTGLGYQTALALAEHGAHVVLAVR
TH_4179|M.thermoresistible__bu MTGKWT---ARDVPDQTGRTAVITGANTGLGFETAKVLAEKGAHVVLAVR
Mflv_0143|M.gilvum_PYR-GCK MT-DWTNWNVADIPDQTGRTAVITGANTGLGFETAKALAEKGARVVIAVR
Mvan_0762|M.vanbaalenii_PYR-1 MT-DWT---TADIPDQTGRTAVITGANTGLGFETAKALAAGGARVVLAVR
MAB_3438|M.abscessus_ATCC_1997 MS-------TYPDRPVAGSRIVITGATNGIGKEIARALVRRGALLTLLAR
MMAR_3998|M.marinum_M ---------------MTGKTIVITGASDGIGAAAARRLTQNGEKVVVVGR
MUL_3862|M.ulcerans_Agy99 ---------------MTGKTIVITGASDGIGAGAARRLTQNGEKVVVVGR
MAV_1610|M.avium_104 ---------------MTRRTIVITGASDGIGAAAARRLSRAGDRIVVVGR
: ::***. *:* * * * :.: *
Mb0069|M.bovis_AF2122/97 NLDKGKQAAARITEAT-PGAEVELQELDLTSLASVRAAAAQLKSDHQRID
Rv0068|M.tuberculosis_H37Rv NLDKGKQAAARITEAT-PGAEVELQELDLTSLASVRAAAAQLKSDHQRID
MSMEG_0876|M.smegmatis_MC2_155 NLDKGKAAVDWIARSA-PTADLELQQLDLGSLASVRAAADDLKGKFDRID
MLBr_00315|M.leprae_Br4923 NLDKGKDAAARITATS-AQNNVALQELDLASLESVRAAAKQLRSDYDHID
TH_4179|M.thermoresistible__bu DPDKGRRAADRITAAA-PHADVTVRQLDLTSLDNIRRAADDLRAGYPRID
Mflv_0143|M.gilvum_PYR-GCK NTDKGAQAAARIRG------DVDVQELDLTSLSSIRTAAEALKARFDKID
Mvan_0762|M.vanbaalenii_PYR-1 DTEKGAQAAAKMAG------DVDVQQLDLTSLASIRSAADALKSRFDHID
MAB_3438|M.abscessus_ATCC_1997 DPAKASATVRELAAENGAITAPEYIQADLADLDSVRAAAHEIATTHPRID
MMAR_3998|M.marinum_M SESKTTAVAAQLRADY--------FVADFADLSQVRALADKIRSEYPRID
MUL_3862|M.ulcerans_Agy99 SESKTTAVAAQLRADY--------FVADFADLSQVRALADKIRSEYPRID
MAV_1610|M.avium_104 SQTKTAAVAAELGADH--------FVVDYADLSQVRALADKMRAQYPRID
. * .. : * .* .:* * : . :**
Mb0069|M.bovis_AF2122/97 LLINNAGVMYTPRQTTADGFEMQFGTNHLGHFALTGLLIDRLLPVAGSRV
Rv0068|M.tuberculosis_H37Rv LLINNAGVMYTPRQTTADGFEMQFGTNHLGHFALTGLLIDRLLPVAGSRV
MSMEG_0876|M.smegmatis_MC2_155 LLVNNAGVMWPPRQTTADGFELQFGTNHLGHFALTGLLLDRMLTVPGSRV
MLBr_00315|M.leprae_Br4923 LLINNAGVMWTPKSTTKDGFELQFGTNHLGHFAFTGLLLDRLLPIVGSRV
TH_4179|M.thermoresistible__bu LLINNAGVMYPPRQTTRDGFELQFGTNHLGHFALTGQLLDNILPVDGSRV
Mflv_0143|M.gilvum_PYR-GCK LLINNAGVMTTPKGTTADGFELQFGTNHLGHFALTGLLFDNILDIPGSRI
Mvan_0762|M.vanbaalenii_PYR-1 LLINNAGVMTTPKGTTADGFELQFGTNHLGHFAFTGLLLDKLLDVDGARI
MAB_3438|M.abscessus_ATCC_1997 TLVNNAGIHSLSGRTSADGFDLMTATNHLGPFLLTNLLLEPIVAAERARI
MMAR_3998|M.marinum_M VLANNAGAMCHEIQFTADGYEKTYQVNYLSPFLLTTLLLDVLVQS-RATV
MUL_3862|M.ulcerans_Agy99 VLANNAGAMCHEFQLTADAYEKTYQVNYLSSFLLTTLRLDVLVQS-RATV
MAV_1610|M.avium_104 VLLNNAGGVASRIELTADGYERTYQVNYLAPFLLTTQLLDVLLES-RATV
* **** : *.:: .*:*. * :* :: :: : :
Mb0069|M.bovis_AF2122/97 VTISSVGHRIRAAIHFDDLQWERRYRRVA---AYGQAKLANLLFTYELQR
Rv0068|M.tuberculosis_H37Rv VTISSVGHRIRAAIHFDDLQWERRYRRVA---AYGQAKLANLLFTYELQR
MSMEG_0876|M.smegmatis_MC2_155 VTVSSQGHRILAAIHFDDLQWERRYNRVA---AYGQSKLANLLFTYELQR
MLBr_00315|M.leprae_Br4923 ITVSSLSHRLFADIHFNDLQWECNYNRVA---AYGQSKLANLLFTYELQR
TH_4179|M.thermoresistible__bu VTVASIAHRNMADIHFDDLQWERGYHRVA---AYGQSKLANLMFAYELQR
Mflv_0143|M.gilvum_PYR-GCK VTVSSNGHKMGGAIHWDDLQWERSYNRMG---AYTQSKLANLLFTYELQR
Mvan_0762|M.vanbaalenii_PYR-1 VTVSSNGHKMGGAIHWDDLQWERSYSRMG---AYSQSKLANLLFTYELQR
MAB_3438|M.abscessus_ATCC_1997 VITASEAHRSWPRIDLERFAEARRYNAIRSEIRYGQSKLMNILFTQELSR
MMAR_3998|M.marinum_M VNTTSSSQRLLPKVTLAELESTAQRRPSV---AYALTKLAIVLFTRELHR
MUL_3862|M.ulcerans_Agy99 VNTTSSSQRLLPKVTLAELENTAQRRPSV---AYALTKLAIVLFTRELHR
MAV_1610|M.avium_104 VNTTSSSHKLILRATVDDLENTANRRPAV---AYAYSKLAIVLFTKELHR
: :* .:: : * :** ::*: ** *
Mb0069|M.bovis_AF2122/97 RLAP-GGTTIAVASHPGVSNTELVRNMPRPLV-AVAAILAP-LMQDAELG
Rv0068|M.tuberculosis_H37Rv RLAP-GGTTIAVASHPGVSNTEVVRNMPRPLV-AVAAILAP-LMQDAELG
MSMEG_0876|M.smegmatis_MC2_155 RLT--GHQTTALAAHPGASNTELARHLPG----ALERLVTP-LAQDAALG
MLBr_00315|M.leprae_Br4923 RLAT-RQTTIAVAAHPGGSRTELTRTLPALIA-PIFSVAELFLTQDAATG
TH_4179|M.thermoresistible__bu RLSAKNAPTISVAAHPGVSNTELTRYIPGARL-PGVSLLAGLLTNSPAVG
Mflv_0143|M.gilvum_PYR-GCK RLAP-RGKTIAVAAHPGTSTTELARNLPRPVERAFLAAAPVLFAQTADRG
Mvan_0762|M.vanbaalenii_PYR-1 RLAP-RGKTIAVAAHPGTSSTELGRNLPAALQPALNRLAP-LLAQSPAAG
MAB_3438|M.abscessus_ATCC_1997 RLDG--TGVTANCFCPGMVSTGLVRDSRVLTAVAGLASRTP-FVRRPDQG
MMAR_3998|M.marinum_M RYHAKGLSAATFHPGYVDSNFGEASGSRALAFMKYHVPLVARLTSTAEQG
MUL_3862|M.ulcerans_Agy99 RYHAKGLSAATFHPGYVDSNFGEASGSRALAFMKYHVPLVTRFTSTAEQG
MAV_1610|M.avium_104 RYHARGLSVAAVHPGNVNSNIGIASGSRFLVFMQRYTP-AALFISSPDQG
* . : : . *
Mb0069|M.bovis_AF2122/97 ALPTLRAATDPAVRG--GQYFGPDGFGEIRGYPKVVASSAQSHDEQLQRR
Rv0068|M.tuberculosis_H37Rv ALPTLRAATDPAVRG--GQYFGPDGFGEIRGYPKVVASSAQSHDEQLQRR
MSMEG_0876|M.smegmatis_MC2_155 ALPTLRAATDPGALG--GQYFGPDGIGETRGYPKVVASSAQSHDADLQRR
MLBr_00315|M.leprae_Br4923 ALPTLRAATDAAVLG--GQYFGPDGFAEIRGHPKVVASNGKSHDVDRQLR
TH_4179|M.thermoresistible__bu ALATLRAATDPEVKG--GQYYGPDGFQEIRGHPVLVGSSAKSRDEDIQRR
Mflv_0143|M.gilvum_PYR-GCK ALPTLRAAADPGVLG--GQYYGPDGLGQQRGAPVVVASSAQSYDVDQQRR
Mvan_0762|M.vanbaalenii_PYR-1 ALPTLRAATDPSVLG--GQYYGPDGIGQQRGNPVVVASSNQSYDMALQRR
MAB_3438|M.abscessus_ATCC_1997 ARMGLRLVLDEDLATTTGQFFTS-----TPGLGQLPAVRIRS-DQKAQEE
MMAR_3998|M.marinum_M AEQLVWLASSAAGTD--------WTSGEYYSRHKIAKANRAAYDPRLASE
MUL_3862|M.ulcerans_Agy99 AEQLVWLASSAAGTD--------WTSGEYYSRHKIARANRAAYDPRLASE
MAV_1610|M.avium_104 ADPLVRLASSPPDSE--------WTSGAYYAKRKIGKTTRLADDPRLAAE
* : . . . : : * .
Mb0069|M.bovis_AF2122/97 LWAVSEELTGVVYPVG-----------------------
Rv0068|M.tuberculosis_H37Rv LWAVSEELTGVVYPVG-----------------------
MSMEG_0876|M.smegmatis_MC2_155 LWAVSEELTGVTFPVGEALQQA-----------------
MLBr_00315|M.leprae_Br4923 LWAVSEELTGVVYPVG-----------------------
TH_4179|M.thermoresistible__bu LWTVSEELTGVTFPV------------------------
Mflv_0143|M.gilvum_PYR-GCK LWEISEELTKVTFPDL-----------------------
Mvan_0762|M.vanbaalenii_PYR-1 LWTVSEELTGVTFPVPVLS--------------------
MAB_3438|M.abscessus_ATCC_1997 LWRRSGELVSL----------------------------
MMAR_3998|M.marinum_M LWDRTLAKLR-----------------------------
MUL_3862|M.ulcerans_Agy99 LWDRTLAKLRREARPATSGIHQIVKSIMPLTMSPLSRSV
MAV_1610|M.avium_104 LWERTAARLG-----------------------------
** :