For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAQQTQVTEEQARALAEESRETGWDKPSFAKELFLGRFPLELIHPFPQPSDADEARTRAFLDQLRKFLDT VDGSVIERDAEIPAEYVRGLADLGCFGMKIPSEFGGLNMSQVAYNRALMMISSVHPSLGALLSAHQSIGV PEPLKLAGTPEQKRKFLPRCAAGAISAFLLTEPDVGSDPARLASTATPVDDGKAYELNGVKLWTTNGVVA ELLVVMARVPKSEGHRGGISAFVVEADSPGITVERRNKFMGLRGIENGVTRLHRVRVPSENLVGREGDGL KIALTTLNAGRLSIPASATGSAKWALKIAREWSGERVQWGKPLAEHEAVARKLSFIAATVYALDAVLELS AQMADEGRNDIRIEAALAKLWSSEMACVIADEVMQIRGGRGYETAESLAARGERAVPVEQVLRDLRINRI FEGASEIMRLLIAREAVDAHLAAAGDLAKPDAGLREKAAAAVGASGFYAKWLPQLVFGEGQRPRAYHEFG ALARHLRFIERSSRKLARNTFYGMARWQAKLEQRQGFLGRVVDIGAELFAMSAACVRAEAQRNADPVLGQ QAYELAEAFCAHATLRVEALFQALWDNTDTGDVALTRNLQRGRYAWLEDGIIDQSEGSGPWIAHWEEGPS TEANLARRFLTI
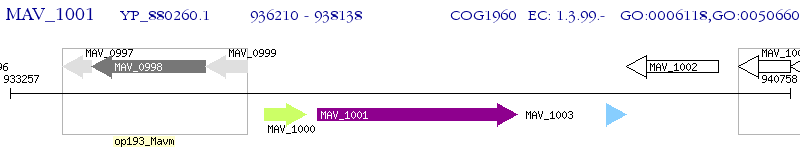
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1001 | - | - | 100% (642) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_4958 | - | 9e-44 | 31.61% (386) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_1571 | - | 2e-43 | 33.07% (387) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0897 | fadE10 | 0.0 | 85.51% (642) | acyl-CoA dehydrogenase FADE10 |
| M. gilvum PYR-GCK | Mflv_4370 | - | 7e-45 | 33.59% (393) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv0873 | fadE10 | 0.0 | 85.51% (642) | acyl-CoA dehydrogenase FADE10 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 8e-43 | 30.56% (360) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1188c | - | 2e-41 | 31.79% (368) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_4660 | fadE10 | 0.0 | 86.25% (640) | acyl-CoA dehydrogenase FadE10 |
| M. smegmatis MC2 155 | MSMEG_2205 | - | 6e-45 | 34.11% (387) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4603 | - | 7e-48 | 33.33% (378) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_0278 | fadE10 | 0.0 | 85.94% (640) | acyl-CoA dehydrogenase FadE10 |
| M. vanbaalenii PYR-1 | Mvan_1476 | - | 2e-45 | 32.64% (386) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4660|M.marinum_M ----MAQQAQVTEEQARALAEESRESGWNKPSFAKELFLGRFPLELINPF
MUL_0278|M.ulcerans_Agy99 ----MAQQAQVTEEQARALAEESRESGWNKPSFAKELFLGRFPLELINPF
Mb0897|M.bovis_AF2122/97 ----MAQQTQVTEEQARALAEESRESGWDKPSFAKELFLGRFPLGLIHPF
Rv0873|M.tuberculosis_H37Rv ----MAQQTQVTEEQARALAEESRESGWDKPSFAKELFLGRFPLGLIHPF
MAV_1001|M.avium_104 ----MAQQTQVTEEQARALAEESRETGWDKPSFAKELFLGRFPLELIHPF
Mflv_4370|M.gilvum_PYR-GCK MASPPANAAEAGIPSAEVNDEDFQAILAATREFVRTAVVPRE--------
MSMEG_2205|M.smegmatis_MC2_155 MTG----TVTDGQSAAEVGDEDFEEILAQARSFIRNAVVPRE--------
TH_4603|M.thermoresistible__bu -----------MAVDRLLPSQEAAELIELTREIADKVLDPIV--------
Mvan_1476|M.vanbaalenii_PYR-1 -----------MAVDRLLPTDEARELVDLARQIADKALDPIV--------
MLBr_00737|M.leprae_Br4923 ----MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHA--------
MAB_1188c|M.abscessus_ATCC_199 ---------------MFNLTEEQREIWNTAREFADIHIAPHA--------
:: : .
MMAR_4660|M.marinum_M PRPSDAEEARTAAFLCRLQEFLGTIDGSVIERDAQIPDEYVNGLAELGCF
MUL_0278|M.ulcerans_Agy99 PRPSDAEEARTAAFLCRLQEFLGTIDGSVIERDAQIPDEYVKGLAELGCF
Mb0897|M.bovis_AF2122/97 PKPSDAEEARTEAFLVKLREFLDTVDGSVIERAAQIPDEYVKGLAELGCF
Rv0873|M.tuberculosis_H37Rv PKPSDAEEARTEAFLVKLREFLDTVDGSVIERAAQIPDEYVKGLAELGCF
MAV_1001|M.avium_104 PQPSDADEARTRAFLDQLRKFLDTVDGSVIERDAEIPAEYVRGLADLGCF
Mflv_4370|M.gilvum_PYR-GCK ---------------------------QEILSTDQVPDDLREQTKSMGLF
MSMEG_2205|M.smegmatis_MC2_155 ---------------------------NEILATDKVPDDIREQAKNMGLF
TH_4603|M.thermoresistible__bu ---------------------------DRHEKDETYPEGVFEQLGAAGLL
Mvan_1476|M.vanbaalenii_PYR-1 ---------------------------DQHEKDETYPDGVFATLGEAGLL
MLBr_00737|M.leprae_Br4923 ---------------------------ADVDQRARFPEEALAALNASGFN
MAB_1188c|M.abscessus_ATCC_199 ---------------------------LDWDRDKYFPVEIFPKAAALGMG
* *
MMAR_4660|M.marinum_M GMKIPSEYGGLSMSQVAYNRALMLASTVHTSLGALLSAHQSIGVPEPLKL
MUL_0278|M.ulcerans_Agy99 GMKIPSEYGGLSMSQVAYNRALMLASTVHTSLGALLSAHQSIGVPEPLKL
Mb0897|M.bovis_AF2122/97 GLKIPSEYGGLNMSQVAYNRVLMMVTTVHSSLGALLSAHQSIGVPEPLKL
Rv0873|M.tuberculosis_H37Rv GLKIPSEYGGLNMSQVAYNRVLMMVTTVHSSLGALLSAHQSIGVPEPLKL
MAV_1001|M.avium_104 GMKIPSEFGGLNMSQVAYNRALMMISSVHPSLGALLSAHQSIGVPEPLKL
Mflv_4370|M.gilvum_PYR-GCK GYAIPQEWGGLGLNLAQDVELAMELGYTSLALRSMFGTNNGIAGQVLVGY
MSMEG_2205|M.smegmatis_MC2_155 GYAIPQQWGGLGLNLAQDVELAMEFGYTSLALRSMFGTNNGIAGQVLVGF
TH_4603|M.thermoresistible__bu SLPQPEEWGGGGQPYEVYLQVLEEIAARWASVAVAVSVHS-LSSHPLLVF
Mvan_1476|M.vanbaalenii_PYR-1 SLPYPEEWGGGGQPYEVYLQVLEELAARWAAVAVAVSVHS-LSCHPLMSF
MLBr_00737|M.leprae_Br4923 AIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVNK-LGTMGLILR
MAB_1188c|M.abscessus_ATCC_199 GIYVNPDVGGSGLTRLDASLIFEAMATGCSAVSAFISIHN-MAAWMIDEF
. : ** . : . :. :.
MMAR_4660|M.marinum_M AGSPEQKKKFLPRCAAGAISAFLLTEPDVGSDPARLASTATPIDGG--QA
MUL_0278|M.ulcerans_Agy99 AGSPEQKKKFLPRCAAGAISAFLLTEPDVGSDPARLASTATPIDDG--QA
Mb0897|M.bovis_AF2122/97 AGTAEQKRRFLPRCAAGAISAFLLTEPDVGSDPARMASTATPIDDG--QA
Rv0873|M.tuberculosis_H37Rv AGTAEQKRRFLPRCAAGAISAFLLTEPDVGSDPARMASTATPIDDG--QA
MAV_1001|M.avium_104 AGTPEQKRKFLPRCAAGAISAFLLTEPDVGSDPARLASTATPVDDG--KA
Mflv_4370|M.gilvum_PYR-GCK GTDEQKKTWLERIASGEIVASFALTEPGAGSNPSGLRTKAVRDGDGPHSG
MSMEG_2205|M.smegmatis_MC2_155 GTDEQKQQWLEGIASGEVVASFALTEPGAGSNPAGLRTKAVRDG----SD
TH_4603|M.thermoresistible__bu GTEEQKKRWLPGMLSGEQIGAYSLSEPQAGSDAAALRCAATPTD----GG
Mvan_1476|M.vanbaalenii_PYR-1 GTDEQRQRWLPEMLGGSTIGAYSLSEPQAGSDAAALTCKAVAAD----GG
MLBr_00737|M.leprae_Br4923 GSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRAKADG----DD
MAB_1188c|M.abscessus_ATCC_199 GDEEQRRRWLPSMCTMETIGAYCLTEPECGSDAAALRTSAIRQG----DE
. :: : :.:: *:* **:.: : * .
MMAR_4660|M.marinum_M YELEGVKLWTTNGVVAELLVVMARVPKSEGHRGGISAFVVEADSPGITVE
MUL_0278|M.ulcerans_Agy99 YELEGIKLWTTNGVVAELLVVMARVPKSEGHRGGISAFVVEADSPGITVE
Mb0897|M.bovis_AF2122/97 YELEGVKLWTTNGVVADLLVVMARVPRSEGHRGGISAFVVEADSPGITVE
Rv0873|M.tuberculosis_H37Rv YELEGVKLWTTNGVVADLLVVMARVPRSEGHRGGISAFVVEADSPGITVE
MAV_1001|M.avium_104 YELNGVKLWTTNGVVAELLVVMARVPKSEGHRGGISAFVVEADSPGITVE
Mflv_4370|M.gilvum_PYR-GCK WILNGQKRFITNAPNADLFVTFARSRPADDKGAGIAVFLVPADTPGVDIG
MSMEG_2205|M.smegmatis_MC2_155 WVINGQKRFITNAPTAGLFVVFARTRPADADGAGIAVFLVPADTPGVEVG
TH_4603|M.thermoresistible__bu YVINGSKSWITHGGKADFYTLFARTG---EGSRGVSCFLVPADQPGLSFG
Mvan_1476|M.vanbaalenii_PYR-1 YRINGAKAWITHGGIADFYNLFARTG---EGSQGISCFLVPRDTEGLTFG
MLBr_00737|M.leprae_Br4923 WILNGFKCWITNGGKSTWYTVMAVTDPD-KGANGISAFIVHKDDEGFSIG
MAB_1188c|M.abscessus_ATCC_199 YVLNGVKQFISGAGAADLYVVMARTG-G-PGPRGISTIVVPKGTPGLSFG
: ::* * : : . : :* *:: ::* . *. .
MMAR_4660|M.marinum_M RRNKFMGLRGIENGVTRLHRVRVPKENLIG-REGDGLRIALTTLNAGRLA
MUL_0278|M.ulcerans_Agy99 RRNKFMGLRGIENGVTRLHRVRVPKENLIG-REGDGLRIALTTLNAGRLA
Mb0897|M.bovis_AF2122/97 RRNKFMGLRGIENGVTRLHRVRVPKDNLIG-REGDGLKIALTTLNAGRLS
Rv0873|M.tuberculosis_H37Rv RRNKFMGLRGIENGVTRLHRVRVPKDNLIG-REGDGLKIALTTLNAGRLS
MAV_1001|M.avium_104 RRNKFMGLRGIENGVTRLHRVRVPSENLVG-REGDGLKIALTTLNAGRLS
Mflv_4370|M.gilvum_PYR-GCK AKDKKMGQEGAWTADVNFTDVEVPDTALVGGSEDLGYRAAMTSLARGRVH
MSMEG_2205|M.smegmatis_MC2_155 PKDAKMGQEGAWTADVNFTDVRVPAAALVGGSEDVGYRAAMTSLARGRVH
TH_4603|M.thermoresistible__bu KPEEKMGLHAVPTTSAFYDNARIDADRRIG-EEGQGLQIAFSALDSGRLG
Mvan_1476|M.vanbaalenii_PYR-1 KPEEKMGLHAVPTTAAHYENAFVEADRRIG-AEGQGLQIAFSALDSGRLG
MLBr_00737|M.leprae_Br4923 PKEKKLGIKGSPTTELYFDKCRIPGDRIIG-EPGTGFKTALATLDHTRPT
MAB_1188c|M.abscessus_ATCC_199 APERKMGWHAQPTAQVVFEDVRVPVANRIG-EEGIGFTIAMRGLNGGRLN
: :* .. . : :* . * *: * *
MMAR_4660|M.marinum_M IPAMVTGAAKWSLKIAREWSGERVQWGKPLGKHEAVAGKISFIAATTYAL
MUL_0278|M.ulcerans_Agy99 IPAMVTGAAKWSLKIAREWSGERVQWGKPLGKDEAVAGKISFIAATTYAL
Mb0897|M.bovis_AF2122/97 LPAIATGVAKQALKIAREWSVERVQWGKPVGQHEAVASKISFIAATNYAL
Rv0873|M.tuberculosis_H37Rv LPAIATGVAKQALKIAREWSVERVQWGKPVGQHEAVASKISFIAATNYAL
MAV_1001|M.avium_104 IPASATGSAKWALKIAREWSGERVQWGKPLAEHEAVARKLSFIAATVYAL
Mflv_4370|M.gilvum_PYR-GCK IAALAVGAAQRALDESVAYAATATQGGTPIGDFQLVQAMLADQQTGVMAG
MSMEG_2205|M.smegmatis_MC2_155 IAALAVGTAQRALDESVAYAATATQGGQPIGNFQLVQAMIADQQTGVMAG
TH_4603|M.thermoresistible__bu IAAVATGLAQAALDEAVAYANERTAFGRKIIDHQGLGFLLADMAAAVATA
Mvan_1476|M.vanbaalenii_PYR-1 IAAVAVGLAQAALDAAVDYSQERTTFGRKIIDHQGLAFLLADMAAAVDAA
MLBr_00737|M.leprae_Br4923 IGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLADMAMKVEAA
MAB_1188c|M.abscessus_ATCC_199 IASCSLGGARSALEKVIEYLRTRKAFGEELIKFQALQFRLADMATELEAA
: : * *: :*. : * : : : :: :
MMAR_4660|M.marinum_M EAVVEISGQMADEGRNDIRIEAALAKLWSSEMACQVADELLQIRGGRGYE
MUL_0278|M.ulcerans_Agy99 EAVVEISGQMADEGRNDIRIEAALAKLWSSEMACQVADELLQIRGGRGYE
Mb0897|M.bovis_AF2122/97 DAVVELSSQMADEGRNDIRIEAALAKLWSSEMACLVGDELLQIRGGRGYE
Rv0873|M.tuberculosis_H37Rv DAVVELSSQMADEGRNDIRIEAALAKLWSSEMACLVGDELLQIRGGRGYE
MAV_1001|M.avium_104 DAVLELSAQMADEGRNDIRIEAALAKLWSSEMACVIADEVMQIRGGRGYE
Mflv_4370|M.gilvum_PYR-GCK RALVRDAARLWVTGQD-RRIAPSAAKLFCTEMAGKVADLAVQIHGGSGYM
MSMEG_2205|M.smegmatis_MC2_155 RALVRDAAQKWVTGED-RRIAPSAAKLYCTEMAGTVADLAVQIHGGTGYM
TH_4603|M.thermoresistible__bu RATYLDAARRRDQGRP-YSQQASIAKLTATDAAMKVTTDAVQVFGGVGYT
Mvan_1476|M.vanbaalenii_PYR-1 RATYLDAARRRDAGLP-YSRHASVAKLTATDAAMKVTTDAVQVFGGAGYT
MLBr_00737|M.leprae_Br4923 RLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTDAVQLFGGAGYT
MAB_1188c|M.abscessus_ATCC_199 RTMVWRAAAAVTEGDPRAAELCAMAKRIATDVGFTVANEALQLHGGYGYL
:. * : :* .:: . : :*: ** **
MMAR_4660|M.marinum_M TAESLAARGERAAPVEQSLRDVRINRIFEGSSEIMRLLIAREAVDAHLTA
MUL_0278|M.ulcerans_Agy99 TAESLAARGERAAPVEQSLRDVRINRIFEGSSEIMRLLIAREAVDAHLTA
Mb0897|M.bovis_AF2122/97 TAESLAARGERAVPVEQMVRDLRINRIFEGSSEIMRLLIAREAVDAHLTA
Rv0873|M.tuberculosis_H37Rv TAESLAARGERAVPVEQMVRDLRINRIFEGSSEIMRLLIAREAVDAHLTA
MAV_1001|M.avium_104 TAESLAARGERAVPVEQVLRDLRINRIFEGASEIMRLLIAREAVDAHLAA
Mflv_4370|M.gilvum_PYR-GCK ----------REVPVERIYRDVRLLRLYEGTSEIQKLIIGGGLVKGAKGK
MSMEG_2205|M.smegmatis_MC2_155 ----------REVPVERIYREVRLLRLYEGTSEIQRLIIGGGLVKAAQRQ
TH_4603|M.thermoresistible__bu ----------RDYRVERYMREAKIMQIFEGTNQIQRLVIARGLTR-----
Mvan_1476|M.vanbaalenii_PYR-1 ----------RDYRVERYMREAKITQIFEGTNQIQRLVISRHLARD----
MLBr_00737|M.leprae_Br4923 ----------SDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR-----
MAB_1188c|M.abscessus_ATCC_199 ----------AEYGIEKIVRDLRVHQILEGTNEIMRVIVSRKLIEQGLPA
:*: *: :: :: **:.:* ::::.
MMAR_4660|M.marinum_M AGDLAKPNTDLRKKAAAAVGASGFYAKWLPQLVFGEGQRPKAYHEFGPLA
MUL_0278|M.ulcerans_Agy99 AGDLAKPNTDLRKKAAAAVGASGFYAKWLPQLVFGEGQRPKAYHEFGPLA
Mb0897|M.bovis_AF2122/97 AGDLANPKADLRQKAAAAAGASGFYAKWLPKLVFGEGQLPTTYREFGALA
Rv0873|M.tuberculosis_H37Rv AGDLANPKADLRQKAAAAAGASGFYAKWLPKLVFGEGQLPTTYREFGALA
MAV_1001|M.avium_104 AGDLAKPDAGLREKAAAAVGASGFYAKWLPQLVFGEGQRPRAYHEFGALA
Mflv_4370|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2205|M.smegmatis_MC2_155 L-------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_1188c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4660|M.marinum_M AHLRFIERSTRKLARNTFYGMARWQAKLEQKQGFLGRIVDIGAELFAMSA
MUL_0278|M.ulcerans_Agy99 AHLRFIERSTRKLARNTFYGMARWQAKLEQKQGFFGRIVDIGAELFAMSA
Mb0897|M.bovis_AF2122/97 THLRFVERSSRKLARNTFYGMARWQASLEKKQGFLGRIVDIGAELFAISA
Rv0873|M.tuberculosis_H37Rv THLRFVERSSRKLARNTFYGMARWQASLEKKQGFLGRIVDIGAELFAISA
MAV_1001|M.avium_104 RHLRFIERSSRKLARNTFYGMARWQAKLEQRQGFLGRVVDIGAELFAMSA
Mflv_4370|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2205|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_1188c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4660|M.marinum_M VCVRAEAQRVADPIVGRQAYELAEAFCGQATLRVEALFHGLWTNTDVGDV
MUL_0278|M.ulcerans_Agy99 VCVRAEAQRVADPIVGRQAYELAEAFCGQATLRVEALFHGLWTNTDVGDV
Mb0897|M.bovis_AF2122/97 ACVRAEAQRTADPVEGEQAYELAEAFCQQATLRVEALFDALWSNTDSIDV
Rv0873|M.tuberculosis_H37Rv ACVRAEAQRTADPVEGEQAYELAEAFCQQATLRVEALFDALWSNTDSIDV
MAV_1001|M.avium_104 ACVRAEAQRNADPVLGQQAYELAEAFCAHATLRVEALFQALWDNTDTGDV
Mflv_4370|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2205|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_1188c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4660|M.marinum_M RLTNDVLEGRYAWLEDGIIDPSEGTGPWIAHWEPGPSTQPNVARRFLPDS
MUL_0278|M.ulcerans_Agy99 RLTNDVLEGRYAWLEDGIIDPSEGTGPWIAHWEPGPSTQPNVARRFLPDS
Mb0897|M.bovis_AF2122/97 RLANDVLEGRYTWLEQGILDQSEGTGPWIASWEPGPSTEANLARRFLTVS
Rv0873|M.tuberculosis_H37Rv RLANDVLEGRYTWLEQGILDQSEGTGPWIASWEPGPSTEANLARRFLTVS
MAV_1001|M.avium_104 ALTRNLQRGRYAWLEDGIIDQSEGSGPWIAHWEEGPSTEANLARRFLTI-
Mflv_4370|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_2205|M.smegmatis_MC2_155 --------------------------------------------------
TH_4603|M.thermoresistible__bu --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_1188c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4660|M.marinum_M TSAEAKL
MUL_0278|M.ulcerans_Agy99 TSVEAKL
Mb0897|M.bovis_AF2122/97 PSSEAKL
Rv0873|M.tuberculosis_H37Rv PSSEAKL
MAV_1001|M.avium_104 -------
Mflv_4370|M.gilvum_PYR-GCK -------
MSMEG_2205|M.smegmatis_MC2_155 -------
TH_4603|M.thermoresistible__bu -------
Mvan_1476|M.vanbaalenii_PYR-1 -------
MLBr_00737|M.leprae_Br4923 -------
MAB_1188c|M.abscessus_ATCC_199 -------