For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAQQTQVTEEQARALAEESRESGWDKPSFAKELFLGRFPLGLIHPFPKPSDAEEARTEAFLVKLREFLDT VDGSVIERAAQIPDEYVKGLAELGCFGLKIPSEYGGLNMSQVAYNRVLMMVTTVHSSLGALLSAHQSIGV PEPLKLAGTAEQKRRFLPRCAAGAISAFLLTEPDVGSDPARMASTATPIDDGQAYELEGVKLWTTNGVVA DLLVVMARVPRSEGHRGGISAFVVEADSPGITVERRNKFMGLRGIENGVTRLHRVRVPKDNLIGREGDGL KIALTTLNAGRLSLPAIATGVAKQALKIAREWSVERVQWGKPVGQHEAVASKISFIAATNYALDAVVELS SQMADEGRNDIRIEAALAKLWSSEMACLVGDELLQIRGGRGYETAESLAARGERAVPVEQMVRDLRINRI FEGSSEIMRLLIAREAVDAHLTAAGDLANPKADLRQKAAAAAGASGFYAKWLPKLVFGEGQLPTTYREFG ALATHLRFVERSSRKLARNTFYGMARWQASLEKKQGFLGRIVDIGAELFAISAACVRAEAQRTADPVEGE QAYELAEAFCQQATLRVEALFDALWSNTDSIDVRLANDVLEGRYTWLEQGILDQSEGTGPWIASWEPGPS TEANLARRFLTVSPSSEAKL
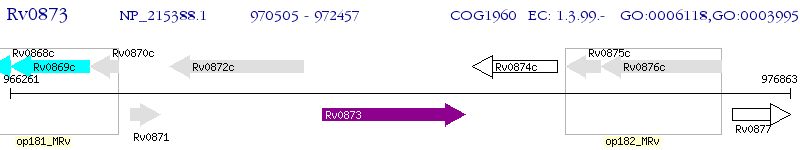
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0873 | fadE10 | - | 100% (650) | PROBABLE ACYL-CoA DEHYDROGENASE FADE10 |
| M. tuberculosis H37Rv | Rv3274c | fadE25 | 5e-44 | 31.54% (390) | acyl-CoA dehydrogenase FADE25 |
| M. tuberculosis H37Rv | Rv2500c | fadE19 | 7e-38 | 31.75% (359) | acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0897 | fadE10 | 0.0 | 100.00% (650) | acyl-CoA dehydrogenase FADE10 |
| M. gilvum PYR-GCK | Mflv_4370 | - | 4e-45 | 33.51% (385) | acyl-CoA dehydrogenase domain-containing protein |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 1e-43 | 31.39% (360) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3618c | - | 7e-41 | 31.67% (360) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_4660 | fadE10 | 0.0 | 85.85% (650) | acyl-CoA dehydrogenase FadE10 |
| M. avium 104 | MAV_1001 | - | 0.0 | 85.51% (642) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2205 | - | 3e-44 | 33.52% (361) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3736 | - | 9e-48 | 33.66% (413) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_0278 | fadE10 | 0.0 | 85.69% (650) | acyl-CoA dehydrogenase FadE10 |
| M. vanbaalenii PYR-1 | Mvan_1476 | - | 1e-44 | 33.70% (359) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0873|M.tuberculosis_H37Rv MAQQTQVTEEQARALAEESRESGWDKPSFAKELFLGRFPLGLIHPFPKPS
Mb0897|M.bovis_AF2122/97 MAQQTQVTEEQARALAEESRESGWDKPSFAKELFLGRFPLGLIHPFPKPS
MMAR_4660|M.marinum_M MAQQAQVTEEQARALAEESRESGWNKPSFAKELFLGRFPLELINPFPRPS
MUL_0278|M.ulcerans_Agy99 MAQQAQVTEEQARALAEESRESGWNKPSFAKELFLGRFPLELINPFPRPS
MAV_1001|M.avium_104 MAQQTQVTEEQARALAEESRETGWDKPSFAKELFLGRFPLELIHPFPQPS
MSMEG_2205|M.smegmatis_MC2_155 ------------------MTGTVTDG---------------QSAAEVGDE
TH_3736|M.thermoresistible__bu ------------------MYDIRRPGGVAIAPPGRTHKEKPMSTAEVSDE
Mflv_4370|M.gilvum_PYR-GCK ------------------MASPPANAAEAG-----------IPSAEVNDE
MLBr_00737|M.leprae_Br4923 ---------------------------------MVGWSGNPLFDLFKLPE
MAB_3618c|M.abscessus_ATCC_199 ------------------------------------MAGNPSFDLFKLGE
Mvan_1476|M.vanbaalenii_PYR-1 ----------------------------------------MAVDRLLPTD
.
Rv0873|M.tuberculosis_H37Rv DAEEARTEAFLVKLREFLDTVDGSVIERAAQIPDEYVKGLAELGCFGLKI
Mb0897|M.bovis_AF2122/97 DAEEARTEAFLVKLREFLDTVDGSVIERAAQIPDEYVKGLAELGCFGLKI
MMAR_4660|M.marinum_M DAEEARTAAFLCRLQEFLGTIDGSVIERDAQIPDEYVNGLAELGCFGMKI
MUL_0278|M.ulcerans_Agy99 DAEEARTAAFLCRLQEFLGTIDGSVIERDAQIPDEYVKGLAELGCFGMKI
MAV_1001|M.avium_104 DADEARTRAFLDQLRKFLDTVDGSVIERDAEIPAEYVRGLADLGCFGMKI
MSMEG_2205|M.smegmatis_MC2_155 DFEEILAQARSFIRNAVVPREN--EILATDKVPDDIREQAKNMGLFGYAI
TH_3736|M.thermoresistible__bu DFRQILRQTREFVRTAVVPREL--EILNEDRVPDDLREQAKQMGLFGYAI
Mflv_4370|M.gilvum_PYR-GCK DFQAILAATREFVRTAVVPREQ--EILSTDQVPDDLREQTKSMGLFGYAI
MLBr_00737|M.leprae_Br4923 EHNELRATIRALAEKEIAPHAA--DVDQRARFPEEALAALNASGFNAIHV
MAB_3618c|M.abscessus_ATCC_199 EHDELRAAIRGLAEKEIAPHAK--DVDEKARFPQEALDALVASGFNAVHV
Mvan_1476|M.vanbaalenii_PYR-1 EARELVDLARQIADKALDPIVD--QHEKDETYPDGVFATLGEAGLLSLPY
: . * * .
Rv0873|M.tuberculosis_H37Rv PSEYGGLNMSQVAYNRVLMMVTTVHSSLGALLSAHQSIGVPEPLKLAGTA
Mb0897|M.bovis_AF2122/97 PSEYGGLNMSQVAYNRVLMMVTTVHSSLGALLSAHQSIGVPEPLKLAGTA
MMAR_4660|M.marinum_M PSEYGGLSMSQVAYNRALMLASTVHTSLGALLSAHQSIGVPEPLKLAGSP
MUL_0278|M.ulcerans_Agy99 PSEYGGLSMSQVAYNRALMLASTVHTSLGALLSAHQSIGVPEPLKLAGSP
MAV_1001|M.avium_104 PSEFGGLNMSQVAYNRALMMISSVHPSLGALLSAHQSIGVPEPLKLAGTP
MSMEG_2205|M.smegmatis_MC2_155 PQQWGGLGLNLAQDVELAMEFGYTSLALRSMFGTNNGIAGQVLVGFGTDE
TH_3736|M.thermoresistible__bu PQQWGGLGLDLAQDVELAMELGYTSLALRSMFGTNNGIAGQVLVGFGTEE
Mflv_4370|M.gilvum_PYR-GCK PQEWGGLGLNLAQDVELAMELGYTSLALRSMFGTNNGIAGQVLVGYGTDE
MLBr_00737|M.leprae_Br4923 PEEYGGQGADSVAACIVIEEVARVDASASLIPAVN-KLGTMGLILRGSEE
MAB_3618c|M.abscessus_ATCC_199 PEEYDGQGADSVAACIVIEEVARVCASSSLIPAVN-KLGTMGLILNGSDE
Mvan_1476|M.vanbaalenii_PYR-1 PEEWGGGGQPYEVYLQVLEELAARWAAVAVAVSVH-SLSCHPLMSFGTDE
*.::.* . : ..: :. : .
Rv0873|M.tuberculosis_H37Rv EQKRRFLPRCAAGAISAFLLTEPDVGSDPARMASTATPIDDG--QAYELE
Mb0897|M.bovis_AF2122/97 EQKRRFLPRCAAGAISAFLLTEPDVGSDPARMASTATPIDDG--QAYELE
MMAR_4660|M.marinum_M EQKKKFLPRCAAGAISAFLLTEPDVGSDPARLASTATPIDGG--QAYELE
MUL_0278|M.ulcerans_Agy99 EQKKKFLPRCAAGAISAFLLTEPDVGSDPARLASTATPIDDG--QAYELE
MAV_1001|M.avium_104 EQKRKFLPRCAAGAISAFLLTEPDVGSDPARLASTATPVDDG--KAYELN
MSMEG_2205|M.smegmatis_MC2_155 QKQQWLEGIASGEVVASFALTEPGAGSNPAGLRTKAVRDGS----DWVIN
TH_3736|M.thermoresistible__bu QKARWLEPIASGEVVASFALTEPGAGSNPAGLRSRAVRDGSGADADWIIN
Mflv_4370|M.gilvum_PYR-GCK QKKTWLERIASGEIVASFALTEPGAGSNPSGLRTKAVRDGDGPHSGWILN
MLBr_00737|M.leprae_Br4923 LKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRAKADGD----DWILN
MAB_3618c|M.abscessus_ATCC_199 LKKQVLPAIASGEAMASYALSEREAGSDAASMRTRAKADGD----DWIIN
Mvan_1476|M.vanbaalenii_PYR-1 QRQRWLPEMLGGSTIGAYSLSEPQAGSDAAALTCKAVAADG----GYRIN
: : . :.:: *:* .**:.: : * .. : ::
Rv0873|M.tuberculosis_H37Rv GVKLWTTNGVVADLLVVMARVPRSEGHRGGISAFVVEADSPGITVERRNK
Mb0897|M.bovis_AF2122/97 GVKLWTTNGVVADLLVVMARVPRSEGHRGGISAFVVEADSPGITVERRNK
MMAR_4660|M.marinum_M GVKLWTTNGVVAELLVVMARVPKSEGHRGGISAFVVEADSPGITVERRNK
MUL_0278|M.ulcerans_Agy99 GIKLWTTNGVVAELLVVMARVPKSEGHRGGISAFVVEADSPGITVERRNK
MAV_1001|M.avium_104 GVKLWTTNGVVAELLVVMARVPKSEGHRGGISAFVVEADSPGITVERRNK
MSMEG_2205|M.smegmatis_MC2_155 GQKRFITNAPTAGLFVVFARTRPADADGAGIAVFLVPADTPGVEVGPKDA
TH_3736|M.thermoresistible__bu GQKRFITNAPVADLFVVFARSRPADERGAGIAVFLVPADAPGVEVGAKDA
Mflv_4370|M.gilvum_PYR-GCK GQKRFITNAPNADLFVTFARSRPADDKGAGIAVFLVPADTPGVDIGAKDK
MLBr_00737|M.leprae_Br4923 GFKCWITNGGKSTWYTVMAVTDPD-KGANGISAFIVHKDDEGFSIGPKEK
MAB_3618c|M.abscessus_ATCC_199 GSKCWITNGGKSSWYTVMAVTDPE-KKANGISAFMVHKDDEGFTVGPLEH
Mvan_1476|M.vanbaalenii_PYR-1 GAKAWITHGGIADFYNLFARTG---EGSQGISCFLVPRDTEGLTFGKPEE
* * : *:. : :* **: *:* * *. . :
Rv0873|M.tuberculosis_H37Rv FMGLRGIENGVTRLHRVRVPKDNLIG-REGDGLKIALTTLNAGRLSLPAI
Mb0897|M.bovis_AF2122/97 FMGLRGIENGVTRLHRVRVPKDNLIG-REGDGLKIALTTLNAGRLSLPAI
MMAR_4660|M.marinum_M FMGLRGIENGVTRLHRVRVPKENLIG-REGDGLRIALTTLNAGRLAIPAM
MUL_0278|M.ulcerans_Agy99 FMGLRGIENGVTRLHRVRVPKENLIG-REGDGLRIALTTLNAGRLAIPAM
MAV_1001|M.avium_104 FMGLRGIENGVTRLHRVRVPSENLVG-REGDGLKIALTTLNAGRLSIPAS
MSMEG_2205|M.smegmatis_MC2_155 KMGQEGAWTADVNFTDVRVPAAALVGGSEDVGYRAAMTSLARGRVHIAAL
TH_3736|M.thermoresistible__bu KMGQEGAWTADVTFTDVRVPASALVGGEEDIGYRAAMTSLARGRVHIAAL
Mflv_4370|M.gilvum_PYR-GCK KMGQEGAWTADVNFTDVEVPDTALVGGSEDLGYRAAMTSLARGRVHIAAL
MLBr_00737|M.leprae_Br4923 KLGIKGSPTTELYFDKCRIPGDRIIG-EPGTGFKTALATLDHTRPTIGAQ
MAB_3618c|M.abscessus_ATCC_199 KLGIKGSPTAELYFENCRIPGDRIIG-EPGTGFKTALQTLDHTRPTIGAQ
Mvan_1476|M.vanbaalenii_PYR-1 KMGLHAVPTTAAHYENAFVEADRRIG-AEGQGLQIAFSALDSGRLGIAAV
:* .. . : :* . * : *: :* * : *
Rv0873|M.tuberculosis_H37Rv ATGVAKQALKIAREWSVERVQWGKPVGQHEAVASKISFIAATNYALDAVV
Mb0897|M.bovis_AF2122/97 ATGVAKQALKIAREWSVERVQWGKPVGQHEAVASKISFIAATNYALDAVV
MMAR_4660|M.marinum_M VTGAAKWSLKIAREWSGERVQWGKPLGKHEAVAGKISFIAATTYALEAVV
MUL_0278|M.ulcerans_Agy99 VTGAAKWSLKIAREWSGERVQWGKPLGKDEAVAGKISFIAATTYALEAVV
MAV_1001|M.avium_104 ATGSAKWALKIAREWSGERVQWGKPLAEHEAVARKLSFIAATVYALDAVL
MSMEG_2205|M.smegmatis_MC2_155 AVGTAQRALDESVAYAATATQGGQPIGNFQLVQAMIADQQTGVMAGRALV
TH_3736|M.thermoresistible__bu AVGTAQRALDESVAYAASATQGGTPIGQFQLVQAMLADQQTGVMAGRALV
Mflv_4370|M.gilvum_PYR-GCK AVGAAQRALDESVAYAATATQGGTPIGDFQLVQAMLADQQTGVMAGRALV
MLBr_00737|M.leprae_Br4923 AVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLADMAMKVEAARLIV
MAB_3618c|M.abscessus_ATCC_199 ALGIAQGALDQAIAYTKDRKQFGKSISDFQGVQFMLADMAMKVEAARLMV
Mvan_1476|M.vanbaalenii_PYR-1 AVGLAQAALDAAVDYSQERTTFGRKIIDHQGLAFLLADMAAAVDAARATY
. * *: :*. : :: * : : : :: *
Rv0873|M.tuberculosis_H37Rv ELSSQMADEGRNDIRIEAALAKLWSSEMACLVGDELLQIRGGRGYETAES
Mb0897|M.bovis_AF2122/97 ELSSQMADEGRNDIRIEAALAKLWSSEMACLVGDELLQIRGGRGYETAES
MMAR_4660|M.marinum_M EISGQMADEGRNDIRIEAALAKLWSSEMACQVADELLQIRGGRGYETAES
MUL_0278|M.ulcerans_Agy99 EISGQMADEGRNDIRIEAALAKLWSSEMACQVADELLQIRGGRGYETAES
MAV_1001|M.avium_104 ELSAQMADEGRNDIRIEAALAKLWSSEMACVIADEVMQIRGGRGYETAES
MSMEG_2205|M.smegmatis_MC2_155 RDAAQKWVTGED-RRIAPSAAKLYCTEMAGTVADLAVQIHGGTGYM----
TH_3736|M.thermoresistible__bu RDAARKWVSGED-RRVAPSAAKLFCTEMAGQVADLAVQIHGGTGYM----
Mflv_4370|M.gilvum_PYR-GCK RDAARLWVTGQD-RRIAPSAAKLFCTEMAGKVADLAVQIHGGSGYM----
MLBr_00737|M.leprae_Br4923 YAAAARAERGEPDLGFISAASKCFASDIAMEVTTDAVQLFGGAGYT----
MAB_3618c|M.abscessus_ATCC_199 YTAAARAERGEKNLGFISSASKCFASDVAMEVTTDAVQLFGGAGYT----
Mvan_1476|M.vanbaalenii_PYR-1 LDAARRRDAGLP-YSRHASVAKLTATDAAMKVTTDAVQVFGGAGYT----
:. * .: :* .:: * : :*: ** **
Rv0873|M.tuberculosis_H37Rv LAARGERAVPVEQMVRDLRINRIFEGSSEIMRLLIAREAVDAHLTAAGDL
Mb0897|M.bovis_AF2122/97 LAARGERAVPVEQMVRDLRINRIFEGSSEIMRLLIAREAVDAHLTAAGDL
MMAR_4660|M.marinum_M LAARGERAAPVEQSLRDVRINRIFEGSSEIMRLLIAREAVDAHLTAAGDL
MUL_0278|M.ulcerans_Agy99 LAARGERAAPVEQSLRDVRINRIFEGSSEIMRLLIAREAVDAHLTAAGDL
MAV_1001|M.avium_104 LAARGERAVPVEQVLRDLRINRIFEGASEIMRLLIAREAVDAHLAAAGDL
MSMEG_2205|M.smegmatis_MC2_155 ------REVPVERIYREVRLLRLYEGTSEIQRLIIGGGLVKAAQRQL---
TH_3736|M.thermoresistible__bu ------REVPVERIYRDVRLLRLYEGTSEIQRLIIGGGLVKAAQRQTN--
Mflv_4370|M.gilvum_PYR-GCK ------REVPVERIYRDVRLLRLYEGTSEIQKLIIGGGLVKGAKGK----
MLBr_00737|M.leprae_Br4923 ------SDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR---------
MAB_3618c|M.abscessus_ATCC_199 ------TDFPVERMMRDAKITQIYEGTNQIQRVVMSRALLTS--------
Mvan_1476|M.vanbaalenii_PYR-1 ------RDYRVERYMREAKITQIFEGTNQIQRLVISRHLARD--------
**: *: :: :::**:.:* ::::.
Rv0873|M.tuberculosis_H37Rv ANPKADLRQKAAAAAGASGFYAKWLPKLVFGEGQLPTTYREFGALATHLR
Mb0897|M.bovis_AF2122/97 ANPKADLRQKAAAAAGASGFYAKWLPKLVFGEGQLPTTYREFGALATHLR
MMAR_4660|M.marinum_M AKPNTDLRKKAAAAVGASGFYAKWLPQLVFGEGQRPKAYHEFGPLAAHLR
MUL_0278|M.ulcerans_Agy99 AKPNTDLRKKAAAAVGASGFYAKWLPQLVFGEGQRPKAYHEFGPLAAHLR
MAV_1001|M.avium_104 AKPDAGLREKAAAAVGASGFYAKWLPQLVFGEGQRPRAYHEFGALARHLR
MSMEG_2205|M.smegmatis_MC2_155 --------------------------------------------------
TH_3736|M.thermoresistible__bu --------------------------------------------------
Mflv_4370|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_3618c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv0873|M.tuberculosis_H37Rv FVERSSRKLARNTFYGMARWQASLEKKQGFLGRIVDIGAELFAISAACVR
Mb0897|M.bovis_AF2122/97 FVERSSRKLARNTFYGMARWQASLEKKQGFLGRIVDIGAELFAISAACVR
MMAR_4660|M.marinum_M FIERSTRKLARNTFYGMARWQAKLEQKQGFLGRIVDIGAELFAMSAVCVR
MUL_0278|M.ulcerans_Agy99 FIERSTRKLARNTFYGMARWQAKLEQKQGFFGRIVDIGAELFAMSAVCVR
MAV_1001|M.avium_104 FIERSSRKLARNTFYGMARWQAKLEQRQGFLGRVVDIGAELFAMSAACVR
MSMEG_2205|M.smegmatis_MC2_155 --------------------------------------------------
TH_3736|M.thermoresistible__bu --------------------------------------------------
Mflv_4370|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_3618c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv0873|M.tuberculosis_H37Rv AEAQRTADPVEGEQAYELAEAFCQQATLRVEALFDALWSNTDSIDVRLAN
Mb0897|M.bovis_AF2122/97 AEAQRTADPVEGEQAYELAEAFCQQATLRVEALFDALWSNTDSIDVRLAN
MMAR_4660|M.marinum_M AEAQRVADPIVGRQAYELAEAFCGQATLRVEALFHGLWTNTDVGDVRLTN
MUL_0278|M.ulcerans_Agy99 AEAQRVADPIVGRQAYELAEAFCGQATLRVEALFHGLWTNTDVGDVRLTN
MAV_1001|M.avium_104 AEAQRNADPVLGQQAYELAEAFCAHATLRVEALFQALWDNTDTGDVALTR
MSMEG_2205|M.smegmatis_MC2_155 --------------------------------------------------
TH_3736|M.thermoresistible__bu --------------------------------------------------
Mflv_4370|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_3618c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv0873|M.tuberculosis_H37Rv DVLEGRYTWLEQGILDQSEGTGPWIASWEPGPSTEANLARRFLTVSPSSE
Mb0897|M.bovis_AF2122/97 DVLEGRYTWLEQGILDQSEGTGPWIASWEPGPSTEANLARRFLTVSPSSE
MMAR_4660|M.marinum_M DVLEGRYAWLEDGIIDPSEGTGPWIAHWEPGPSTQPNVARRFLPDSTSAE
MUL_0278|M.ulcerans_Agy99 DVLEGRYAWLEDGIIDPSEGTGPWIAHWEPGPSTQPNVARRFLPDSTSVE
MAV_1001|M.avium_104 NLQRGRYAWLEDGIIDQSEGSGPWIAHWEEGPSTEANLARRFLTI-----
MSMEG_2205|M.smegmatis_MC2_155 --------------------------------------------------
TH_3736|M.thermoresistible__bu --------------------------------------------------
Mflv_4370|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_3618c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv0873|M.tuberculosis_H37Rv AKL
Mb0897|M.bovis_AF2122/97 AKL
MMAR_4660|M.marinum_M AKL
MUL_0278|M.ulcerans_Agy99 AKL
MAV_1001|M.avium_104 ---
MSMEG_2205|M.smegmatis_MC2_155 ---
TH_3736|M.thermoresistible__bu ---
Mflv_4370|M.gilvum_PYR-GCK ---
MLBr_00737|M.leprae_Br4923 ---
MAB_3618c|M.abscessus_ATCC_199 ---
Mvan_1476|M.vanbaalenii_PYR-1 ---