For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSCMANREGQKLFSRSRQMPATPSWFTAALDQKPERCDVEVDGCRIRMRVWGAADQPPVVLVHGGGAHSG WWDHIAPFFSHTHRVIAPDLSGHGDSDPRDSYDLRIWAGEVMAAAAAAGPAGRPTIVGHSMGGWVVSTAA MHYSSKINSMLVIDSPLWERAPEEGRLRRRKHTEYRTRDEILARFTAVPSQELILPYVREHIAAESIRKT SAGWVWKFDPAIFGGEFLQPTPADEESLESMLTAMRCRVGYLRCEAGLVPPAMAERIRSLLQLRGPFVEL AQAGHHPMLDQPLPLVATLRTLLEFWSIT*
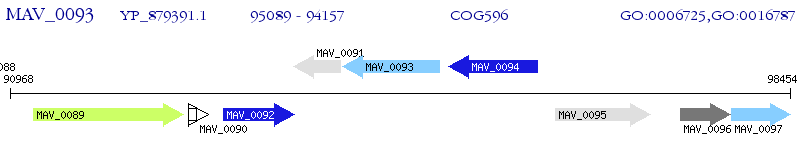
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0093 | - | - | 100% (310) | alpha/beta hydrolase fold |
| M. avium 104 | MAV_1254 | - | 7e-12 | 26.89% (264) | hydrolase, alpha/beta fold family protein |
| M. avium 104 | MAV_2706 | - | 1e-11 | 25.71% (280) | hydrolase, alpha/beta fold family protein, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2734 | - | 3e-11 | 33.62% (116) | hydrolase |
| M. gilvum PYR-GCK | Mflv_0736 | - | 1e-120 | 66.11% (298) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv2715 | - | 3e-11 | 33.62% (116) | hydrolase |
| M. leprae Br4923 | MLBr_02297 | - | 2e-07 | 32.38% (105) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_3034 | - | 9e-12 | 33.08% (133) | hydrolase |
| M. marinum M | MMAR_4837 | - | 3e-11 | 25.26% (194) | hydrolase |
| M. smegmatis MC2 155 | MSMEG_0460 | - | 1e-121 | 66.02% (309) | alpha/beta hydrolase fold |
| M. thermoresistible (build 8) | TH_0684 | hpx | 6e-12 | 37.84% (111) | POSSIBLE NON-HEME HALOPEROXIDASE HPX |
| M. ulcerans Agy99 | MUL_3358 | - | 2e-10 | 32.46% (114) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_2912 | - | 1e-16 | 28.57% (280) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2734|M.bovis_AF2122/97 -----MTERKRNLRPVRD------------------VAPPTLQFRTVHGY
Rv2715|M.tuberculosis_H37Rv -----MTERKRNLRPVRD------------------VAPPTLQFRTVHGY
MUL_3358|M.ulcerans_Agy99 -----MTERKRNLRPVRE------------------VTAPSLQFRTIHGY
MAB_3034|M.abscessus_ATCC_1997 -----MNPPPRLLRPVPDTEHIDGVDGGGDTPSAATSTRPRLAFRTIHGY
Mvan_2912|M.vanbaalenii_PYR-1 -------------------------------------MHDDVKYLDLHGD
MAV_0093|M.avium_104 MLSCMANREGQKLFSRSRQMP-----ATPSWFTAALDQKPERCDVEVDGC
MSMEG_0460|M.smegmatis_MC2_155 ---MRHADEVSPLFQRQRPTPSDPPEGTPEWFTAALEHRPQHSTIDFEGC
Mflv_0736|M.gilvum_PYR-GCK ------------MFSRSRSVD-----EAPAWFTTALADKPEHSHVDVEGC
TH_0684|M.thermoresistible__bu --------------------------------VEQMPPGRVVSVTAPDGV
MLBr_02297|M.leprae_Br4923 -----MAMSNPSVTRIAGP------------WHHLDVHANGIRFHVVEAV
MMAR_4837|M.marinum_M ----MAATETPDSQRLTTS---------------DHMTISQGRIRTSDGI
..
Mb2734|M.bovis_AF2122/97 RRAFR----IAGS--GPAILLIHGIGDNSTTWNGVHAKLA-QRFTVIAPD
Rv2715|M.tuberculosis_H37Rv RRAFR----IAGS--GPAILLIHGIGDNSTTWNGVHAKLA-QRFTVIAPD
MUL_3358|M.ulcerans_Agy99 KRAFR----IAGS--GPAILLIHGIGDNSTTWTGIHAKLA-QRFTVIAPD
MAB_3034|M.abscessus_ATCC_1997 RRAYR----IAGS--GPVLLLIHGIGDNSATWDSVHAQLA-EHFTVIAPD
Mvan_2912|M.vanbaalenii_PYR-1 RVAYR----EAGQ--GPAVLLIHGMGGSSLTWKALLPHLA-TRYRVIAPD
MAV_0093|M.avium_104 RIRMR----VWGAADQPPVVLVHGGGAHSGWWDHIAPFFS-HTHRVIAPD
MSMEG_0460|M.smegmatis_MC2_155 RIHLR----TWGDPENPPLVFVHGGAAHSGWWDHIAPFFT-RTHRVIAPD
Mflv_0736|M.gilvum_PYR-GCK EIHVR----VWGARENPPLVLVHGGAAHSGWWDHIAPLLS-STHRIVAPD
TH_0684|M.thermoresistible__bu RLHAE----VFGPADAPPIVLAHGITCAIRVWAHQIAELA-DDYRVIAFD
MLBr_02297|M.leprae_Br4923 PTQPAGQDCAPSATARPLVMLLHGFGSFWWSWRHQLRGLT-EAR-LVAVD
MMAR_4837|M.marinum_M TLVAD---CYRHAATRPVVLLLHGGGQNRHAWATTARRLHSHGYTVVAYD
* ::: ** * : ::* *
Mb2734|M.bovis_AF2122/97 LLGHGQSDKPRA--DYSVAAYANGMRDLLSVLD--IERVTIVGHSLGGGV
Rv2715|M.tuberculosis_H37Rv LLGHGQSDKPRA--DYSVAAYANGMRDLLSVLD--IERVTIVGHSLGGGV
MUL_3358|M.ulcerans_Agy99 LLGHGRSDKPRA--DYSIAAYANGMRDLLSVLD--IERVTIIGHSLGGGV
MAB_3034|M.abscessus_ATCC_1997 LLGHGQSDKPRA--DYSVAAYANGMRDLLAVLD--IERVTVVGHSLGGGV
Mvan_2912|M.vanbaalenii_PYR-1 LLGHGQSDKPRG--DYSLGAFAVWLRDLLDLLG--IARVTLVGHSLGGGV
MAV_0093|M.avium_104 LSGHGDSD-PRD--SYDLRIWAGEVMAAAAAAGP-AGRPTIVGHSMGGWV
MSMEG_0460|M.smegmatis_MC2_155 VSGHGDSG-TRS--HYSLTKWAREVLAVAAAEGP-AARPTIVGHSMGGWV
Mflv_0736|M.gilvum_PYR-GCK LSGHGDSG-TRD--SYDLRRWSREVMAAAAAFS--DGHPTIVGHSLGGWV
TH_0684|M.thermoresistible__bu HRGHGRSSVPRRRWQYSLDHLASDLDAVLDATLDPGERAVIAGHSMGGIA
MLBr_02297|M.leprae_Br4923 LRGYGGSDKPPR--GYDGWTLAGDTAGLIRALG--HSSATLVGHAEGGLA
MMAR_4837|M.marinum_M TRGHGDSDWDPSG-QYDVERFVSDLISVRDHVSP-DSPPAVVGASLGGLI
*:* *. *. .: * : **
Mb2734|M.bovis_AF2122/97 AMQFAYQFPQLVDR----LILVSA---GGVTKDV-NIVFRLASLPMGSEA
Rv2715|M.tuberculosis_H37Rv AMQFAYQFPQLVDR----LILVSA---GGVTKDV-NIVFRLASLPMGSEA
MUL_3358|M.ulcerans_Agy99 AMQFAYQFPQLVDR----LILVGA---GGVTKDV-NVVFRLASLPMGAEA
MAB_3034|M.abscessus_ATCC_1997 AMQFTYQFPHLVER----LILVAP---GGVTKDV-NIVLRCASLPFIGDA
Mvan_2912|M.vanbaalenii_PYR-1 AMQFVHQHPDYCER----LVLISS---GGLGPEL-GRSLRLLSTPG----
MAV_0093|M.avium_104 VSTAAMHYSSKINS----MLVIDSPLWERAPEEG-RLRRRK--HTEYRTR
MSMEG_0460|M.smegmatis_MC2_155 TATAAMKYGADIDS----ILIIDSPLRERAPEEG-RLRNRRSDHPGYPSR
Mflv_0736|M.gilvum_PYR-GCK TATAASHFGDQINS----IVVVDSPLRDRAPEER-HLSRRT--PKAYRSK
TH_0684|M.thermoresistible__bu ISSWAERYPERVSERAAAVALINTTTGDLLHNVQ-LLPVPDRFAAVRVRT
MLBr_02297|M.leprae_Br4923 CWATALLHPRLVRA----IALINSPHPAALRRSM-LTHRNQGYALLPTLL
MMAR_4837|M.marinum_M ILATHLLAPPDLWAAVVLVDITPRMEFDGAHRIVSFMAAHPDGFGTLDDA
: :
Mb2734|M.bovis_AF2122/97 MALLRLPLVLPAVQIAGRIVGKAIGTTSLG--------HDLPNVLRILDD
Rv2715|M.tuberculosis_H37Rv MALLRLPLVLPAVQIAGRIVGKAIGTTSLG--------HDLPNVLRILDD
MUL_3358|M.ulcerans_Agy99 LALLRLPMVLPTVQLAGKVLGMALGSTALG--------QDLPNVLRILDD
MAB_3034|M.abscessus_ATCC_1997 LGLLRLPMAMPMLRLGGAVARATFGRASMA--------RDIPDVLRVLAD
Mvan_2912|M.vanbaalenii_PYR-1 IELLLPVAAAPSVVAVGERVRSWLGARGLAS-------PEVGETWNAYAS
MAV_0093|M.avium_104 DEILARFTAVPSQELILPYVREHIAAESIR---------------KTSAG
MSMEG_0460|M.smegmatis_MC2_155 EEILARFRAVPAQDVTLPFIDRHIASESVR---------------QTDTG
Mflv_0736|M.gilvum_PYR-GCK DEIVGRFRPVPAQQGVLPYVSAHIASESVR---------------RTLKG
TH_0684|M.thermoresistible__bu ASAVIKTVAGMPARLAAVPSRYVVSMMAVGR-------DADPAITNFVHE
MLBr_02297|M.leprae_Br4923 RYQLPIWPERLLTRNNAAEIERLVRSRSSAKWQACEDFAVTIDHLRIAIQ
MMAR_4837|M.marinum_M ADVIAEYNPRRARPENLDGLHKVLRQRSDGR------------WIWRWDP
: . .
Mb2734|M.bovis_AF2122/97 LPEPTASAAFGRTLRAVVDWRGQMVTMLDRCYLTEAIPVQIIWGTKDVVL
Rv2715|M.tuberculosis_H37Rv LPEPTASAAFGRTLRAVVDWRGQMVTMLDRCYLTEAIPVQIIWGTKDVVL
MUL_3358|M.ulcerans_Agy99 LPEPTASSAFTRTLRAVVDWRGQIVTMLDRCYLTQAIPVQIIWGSRDAVV
MAB_3034|M.abscessus_ATCC_1997 LPEPRASAAFTRTLRAVVDWRGQVVTMLDRCYLTESVPVQLIWGSDDLVI
Mvan_2912|M.vanbaalenii_PYR-1 LSDPQTRTAFLRTLRSVVDAQGQAVSALNRLHFTLGLPTLLIWGDQDRLI
MAV_0093|M.avium_104 WVWKFDPAIFGGEFLQPTPAD---EESLESMLTAMRCRVGYLRCEAGLVP
MSMEG_0460|M.smegmatis_MC2_155 WVWKFDPAIFSGSLQGQTPAE---QEILENTFAQIPCRVGYLRCENGLVP
Mflv_0736|M.gilvum_PYR-GCK WVWKFDPAIFGGHLAEQTPAE---REVMEAMMNQMPCRIGYLRCEHGLVP
TH_0684|M.thermoresistible__bu LFTETPPAARDGWARTLVDAMG----PRHIGLHNLTVPTLVIGSDRDRLL
MLBr_02297|M.leprae_Br4923 IPAAAHCALEYQRWAVRSQLRNEGRKFMKSMSRPFSVPLLHVRGDADPYL
MMAR_4837|M.marinum_M AFISSNFDVLQGNLMTGSEEFDAISGFLADAARRITAPTLLVRGALSDVV
: .
Mb2734|M.bovis_AF2122/97 PVRHAHMAHAAMPGSQLEIFEGSGHFPFHDDPARFIDIVERFMDTTEPAE
Rv2715|M.tuberculosis_H37Rv PVRHAHMAHAAMPGSQLEIFEGSGHFPFHDDPARFIDIVERFMDTTEPAE
MUL_3358|M.ulcerans_Agy99 PVRHAEMAHAAMPGSKLEVFEGSGHFPFHDDPARFIDIVLRFIDSTQPPE
MAB_3034|M.abscessus_ATCC_1997 PVSHGHLAHAAMPGSALEIFDKSGHFPFHDDPERFIGIVRQFIASTEPGE
Mvan_2912|M.vanbaalenii_PYR-1 PPAHGEAAHAALPGSRLVILPAVGHFPQVEAPLAVADTLDDFIANSSP--
MAV_0093|M.avium_104 PAMAERIRSLLQLRGPFVELAQAGHHPMLDQPLPLVATLRTLLEFWSIT-
MSMEG_0460|M.smegmatis_MC2_155 PDMAEQIRSTLQLRGPFVELALAGHHPMLDQPLPLVATIRTLLEFWSIT-
Mflv_0736|M.gilvum_PYR-GCK PEMADVVRDILQLRGPFVDLVEAGHHPMLDQPLPLVATLRTLLEVWSIT-
TH_0684|M.thermoresistible__bu PLVSSRRIAEAAPNLFDFVELPGGHCAILECPDRVNDHLRALAAAAADAR
MLBr_02297|M.leprae_Br4923 LADSVDRTRRYAPHGRYVSVSDAGHFSHEEAPEEINRYLTRFLKQVHGPR
MMAR_4837|M.marinum_M SQDTVNEFLQLVPHAETTDVTGTGHMVAGDNNDAFTAAVTDFLDRTMGMT
** : . : :
Mb2734|M.bovis_AF2122/97 YDQAALRALLRRGGGEATVTGSADTRVAVLNAIGSNERSAT
Rv2715|M.tuberculosis_H37Rv YDQAALRALLRRGGGEATVTGSADTRVAVLNAIGSNERSAT
MUL_3358|M.ulcerans_Agy99 YDQAALRELLRTGGGERTVSDPADTRVAVLSAMGSDERSAT
MAB_3034|M.abscessus_ATCC_1997 YNEDELRRLLRTGVTEHAISGAARTRMAVLDAMGTDERSAT
Mvan_2912|M.vanbaalenii_PYR-1 ----------WHGPPRHADHAEAD-----------------
MAV_0093|M.avium_104 -----------------------------------------
MSMEG_0460|M.smegmatis_MC2_155 -----------------------------------------
Mflv_0736|M.gilvum_PYR-GCK -----------------------------------------
TH_0684|M.thermoresistible__bu RATS-------------------------------------
MLBr_02297|M.leprae_Br4923 LS---------------------------------------
MMAR_4837|M.marinum_M -----------------------------------------