For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ATHDGAQLHVRSYGSAFGQPIVLIHGFGCRIEYWNPQINALAAKYRVIAYDQRGLGRSTLGSDGVRPDVL GRDLAAVLDAVLAPGERAVLVGHSFGGITIMAWAQRFAAHAVSAVLLANTIAERFSATTMLLPFADRYRH IRRPLITALAGSTMTLPSIRWSRWPLQRMALSRYADRQAVDFVYEILADCRTDVRAVWGAGLSGLDVTAG LEKLTVPTTVVTGTEDRLTPPAVAHRIAEALQVRGSLHRFARFPRAGHCTNIEVPYAFNAEIEQLADATR *
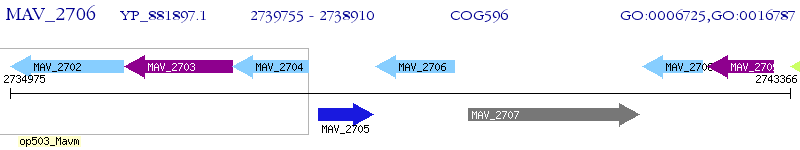
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2706 | - | - | 100% (281) | hydrolase, alpha/beta fold family protein, putative |
| M. avium 104 | MAV_4061 | - | 7e-37 | 40.61% (293) | hydrolase, alpha/beta fold family protein |
| M. avium 104 | MAV_4371 | - | 7e-13 | 27.67% (300) | hydrolase, alpha/beta fold family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3196c | hpx | 6e-36 | 36.18% (293) | non-Heme haloperoxidase Hpx |
| M. gilvum PYR-GCK | Mflv_4502 | - | 2e-38 | 37.67% (292) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3171c | hpx | 9e-36 | 36.18% (293) | non-Heme haloperoxidase Hpx |
| M. leprae Br4923 | MLBr_00862 | ephD | 1e-10 | 22.97% (296) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3738c | - | 4e-16 | 29.57% (301) | putative hydrolase alpha/beta fold |
| M. marinum M | MMAR_1388 | hpx | 2e-32 | 35.17% (290) | non-heme haloperoxidase Hpx |
| M. smegmatis MC2 155 | MSMEG_2036 | - | 5e-38 | 38.68% (287) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_0684 | hpx | 4e-42 | 39.38% (292) | POSSIBLE NON-HEME HALOPEROXIDASE HPX |
| M. ulcerans Agy99 | MUL_2488 | hpx | 1e-32 | 35.17% (290) | non-heme haloperoxidase Hpx |
| M. vanbaalenii PYR-1 | Mvan_1865 | - | 1e-36 | 37.02% (289) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4502|M.gilvum_PYR-GCK ---------------------------MARSSSTRSRRAGSGPAVELPRG
Mvan_1865|M.vanbaalenii_PYR-1 ---------------------------MAPGTSTRSRSRGFAPAQALPPG
TH_0684|M.thermoresistible__bu -------------------------------------------VEQMPPG
MSMEG_2036|M.smegmatis_MC2_155 -----------------------------------------MTLPQLPPG
Mb3196c|M.bovis_AF2122/97 --------------------------------------------------
Rv3171c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1388|M.marinum_M ------------------------MSARKATDGKAHTKAPAALPDVLPPS
MUL_2488|M.ulcerans_Agy99 ------------------------MSARKATDGKAHTKASAALPDVLPPS
MAV_2706|M.avium_104 --------------------------------------------------
MAB_3738c|M.abscessus_ATCC_199 MSNKSNAWLAGVAGLGAVVAVAGVGTARSIGRRRFDDPYRGENFDLLQTD
MLBr_00862|M.leprae_Br4923 ----------------------------------------MPATQSIHKV
Mflv_4502|M.gilvum_PYR-GCK RVVDVRSRDGVRLHAEVFGPEDG-YPIVLAHGITCAIPVWAHQIADLSRD
Mvan_1865|M.vanbaalenii_PYR-1 RIVDVRSKDGVRLHAEVFGPEDG-YPIVLAHGITCSLRVWAYQIAELARD
TH_0684|M.thermoresistible__bu RVVSVTAPDGVRLHAEVFGPADA-PPIVLAHGITCAIRVWAHQIAELADD
MSMEG_2036|M.smegmatis_MC2_155 RTFEVRAVDGVRLHAQTFGPEDG-YPIVFAHGITCAIEVWAHQIADLAGD
Mb3196c|M.bovis_AF2122/97 --MTVRAADGTPLHTQVFGPPHG-YPIVLTHGFVCAIRAWAYQIADLAGD
Rv3171c|M.tuberculosis_H37Rv --MTVRAADGTPLHTQVFGPPHG-YPIVLTHGFVCAIRAWAYQIADLAGD
MMAR_1388|M.marinum_M RSLTVRAADGTALHTQVFGPADG-YPIVLTHGFVCSIRAWAYQIADLATD
MUL_2488|M.ulcerans_Agy99 RSLTVRAADGTALHTQVFGPADG-YPIVLTHGFVCSIRAWAYQIADLATD
MAV_2706|M.avium_104 ----MATHDGAQLHVRSYGSAFG-QPIVLIHGFGCRIEYWNPQINALAAK
MAB_3738c|M.abscessus_ATCC_199 RGSIVTTDDGVPLAVREVGPSNAPLTVVFVHGFCLQMASFHFQRRELASR
MLBr_00862|M.leprae_Br4923 PHQFVESPDGVRLAIYQSGQPEG-PAIVLVHGFPDSHVLWDGVVPLLAKR
: : **. * * . .:*: **: : *:
Mflv_4502|M.gilvum_PYR-GCK H----RVIAYDHRGHGRSGVPPRRGGYSLDYLASDLDAVLEATMAPGERA
Mvan_1865|M.vanbaalenii_PYR-1 H----RVIAFDHRGHGRSAIPPRRGGYSLDYLAGDLDAVLEATLAPGERA
TH_0684|M.thermoresistible__bu Y----RVIAFDHRGHGRSSVPRRRWQYSLDHLASDLDAVLDATLDPGERA
MSMEG_2036|M.smegmatis_MC2_155 Y----RVIAYDHRGHGRSETPRGRNRYSLNHLAADLDAVLDATLRPGERA
Mb3196c|M.bovis_AF2122/97 Y----RVIAFDHRGHGRSGVP-RRGAYSLNHLAADLDSVLDATLAPRERA
Rv3171c|M.tuberculosis_H37Rv Y----RVIAFDHRGHGRSGVP-RRGAYSLNHLAADLDSVLDATLAPRERA
MMAR_1388|M.marinum_M Y----RVIAFDHRGHGRSGIP-RRGGYTLEHLASDLDSVLDATLAPHERA
MUL_2488|M.ulcerans_Agy99 Y----RVIAFDHRGHGRSGIP-RRGGYTLEHLASDLDSVLDATLAPHERA
MAV_2706|M.avium_104 Y----RVIAYDQRGLGRSTLG--SDGVRPDVLGRDLAAVLDAVLAPGERA
MAB_3738c|M.abscessus_ATCC_199 WGDNVRMVFYDQRGHGRSGLP-APASCTVGQLGDDLESVLR-VLVPRGNA
MLBr_00862|M.leprae_Br4923 F----RIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVID-ELSPDRPV
*:: :*:** *:* :. *: :*: : * .
Mflv_4502|M.gilvum_PYR-GCK VIAGHSMGGIAISSWSERLPHRVAERADGVALINTTTGDLLRNVNLLPVP
Mvan_1865|M.vanbaalenii_PYR-1 VIAGHSMGGIAISSWSERLPHRVEQRADAVALINTTTGDLLRNVNLLPVP
TH_0684|M.thermoresistible__bu VIAGHSMGGIAISSWAERYPERVSERAAAVALINTTTGDLLHNVQLLPVP
MSMEG_2036|M.smegmatis_MC2_155 VIAGHSMGGIAITSWAQRYPSRVQTCADGVALINTSTGDLLRDVQLAQVP
Mb3196c|M.bovis_AF2122/97 VVAGHSMGGITIAAWSDRYRHKVRRRTDAVALINTTTGDLVRKVKLLSVP
Rv3171c|M.tuberculosis_H37Rv VVAGHSMGGITIAAWSDRYRHKVRRRTDAVALINTTTGDLVRKVKLLSVP
MMAR_1388|M.marinum_M LIAGHSMGGIAISAWSERYRHKVHRRADAVALINTATGDLLRKIRMLSVP
MUL_2488|M.ulcerans_Agy99 LIAGHSMGGIAISAWSERYRHKVHRRADAVALINTATGDLLRKIRMLSVP
MAV_2706|M.avium_104 VLVGHSFGGITIMAWAQRFAAHAVS---AVLLANTIAERFSATTMLLPFA
MAB_3738c|M.abscessus_ATCC_199 VLVGHSMGGMTVLAHARRHPEQYGRRIVGVGLIASAAEGLSHTAIGEGLR
MLBr_00862|M.leprae_Br4923 HVLAHDWG--SVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSGLR
: .*. * :: .. : .
Mflv_4502|M.gilvum_PYR-GCK -ARFADARVRAAGGVIRTFGGTSMVRVADLPSRRFVSMIAVGRDADPAIA
Mvan_1865|M.vanbaalenii_PYR-1 -PALADVRVRAAGEVLRRFGGVPLVRAVDRPNRRFVSSIAVGRDADPAIA
TH_0684|M.thermoresistible__bu -DRFAAVRVRTASAVIKTVAGMP-ARLAAVPSRYVVSMMAVGRDADPAIT
MSMEG_2036|M.smegmatis_MC2_155 -TRLSTTRIRTAGTILKTFGSTPVPKLADAANRRFVAHLAVGRDADPWVA
Mb3196c|M.bovis_AF2122/97 -RELSPVRVLAGRSLVNTFGGFPLPGAARALSRHVISTLAVAADADPSAT
Rv3171c|M.tuberculosis_H37Rv -RELSPVRVLAGRSLVNTFGGFPLPGAARALSRHVISTLAVAADADPSAT
MMAR_1388|M.marinum_M -RELSAARDFAGRGLINAFGGFPLPSAVRIPTQYLIAMMAVGADAHPTAA
MUL_2488|M.ulcerans_Agy99 -RELSAARDFAGRGLINAFGGFPLPSAVRIPTQYLIAMMAVGADAHPTAA
MAV_2706|M.avium_104 -DRYRHIR----RPLITALAGSTMTLPSIRWSRWPLQRMALSRYADRQAV
MAB_3738c|M.abscessus_ATCC_199 NPALRVLRTAVHYAPGPAHHGRGAIKSLVGP--VLQAASYGGHRVSPTLV
MLBr_00862|M.leprae_Br4923 -RPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMVD
.
Mflv_4502|M.gilvum_PYR-GCK DFVFELFHRTPPAGRGGWARVLVDHL--------GPRHIGLDNLTVPALV
Mvan_1865|M.vanbaalenii_PYR-1 DFVFELFNSTSPAGRGGWARVLVDHL--------GPRHIALKNLTVPTLV
TH_0684|M.thermoresistible__bu NFVHELFTETPPAARDGWARTLVDAM--------GPRHIGLHNLTVPTLV
MSMEG_2036|M.smegmatis_MC2_155 DFVYRLFATTPPTGRGAWGRVLVDNL--------GPKHIPLHNLTVPTLV
Mb3196c|M.bovis_AF2122/97 RLVYELFTQMSAAGRGGCAKMLVEEV--------GSAHLNLDGLTVPTLV
Rv3171c|M.tuberculosis_H37Rv RLVYELFTQTSAAGRGGCAKMLVEEV--------GSAHLNLDGLTVPTLV
MMAR_1388|M.marinum_M RLLYELFAQTSAAGRGGCARMLVGEV--------GSRHISLDGLTVPTLV
MUL_2488|M.ulcerans_Agy99 RLLYELFAQTSAAGRGGCARMLVGEV--------GSRHISLDGLTVPTLV
MAV_2706|M.avium_104 DFVYEILADCRTDVRAVWGAGLSG----------LDVTAGLEKLTVPTTV
MAB_3738c|M.abscessus_ATCC_199 KFSERMIHQTPVTTIVDFLRALEQHD--------ETAALPTIAP-LPSLV
MLBr_00862|M.leprae_Br4923 NISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQL
: : :* :
Mflv_4502|M.gilvum_PYR-GCK IGSTHDRLLPVSSARHIASAAPN---LARFVELA-GGHCAILERPEEVNQ
Mvan_1865|M.vanbaalenii_PYR-1 IGSTKDRLLPMASARRIAATAPN---LARFVEVP-GGHCAILERPDDVND
TH_0684|M.thermoresistible__bu IGSDRDRLLPLVSSRRIAEAAPN---LFDFVELP-GGHCAILECPDRVND
MSMEG_2036|M.smegmatis_MC2_155 IGSAQDRLLPINASRRIADTVPN---LAAFVELR-GGHCGILERPDEVNA
Mb3196c|M.bovis_AF2122/97 IGGVRDRLTPISQSRRIARTAPN---VVGLVELP-GGHCSMLERHQEVNS
Rv3171c|M.tuberculosis_H37Rv IGGVRDRLTPISQSRRIARTAPN---VVGLVELP-GGHCSMLERHQEVNS
MMAR_1388|M.marinum_M IGSERDRLTPISQTRKIASTAPN---VFDLVELP-GGHCSMLEQHHEVNR
MUL_2488|M.ulcerans_Agy99 IGSERDRLTPISQTRKIASTAPN---VFDLVELP-GGHCSMLEQHHEVNR
MAV_2706|M.avium_104 VTGTEDRLTPPAVAHRIAEALQVRGSLHRFARFPRAGHCTNIEVPYAFNA
MAB_3738c|M.abscessus_ATCC_199 ICGDTDMLTPHTQSESMAARLAASG-HNELILVRESGHLVQLEHPEIVND
MLBr_00862|M.leprae_Br4923 IVNTMDRYVRPYGYDATARWVPR-----LWRRDIKAGHFSPMSHPQVMAA
: . * * .** :. .
Mflv_4502|M.gilvum_PYR-GCK QLRWLVDSVSRSTRASAATGDQARASEATGDQARASAATGDQARASSR--
Mvan_1865|M.vanbaalenii_PYR-1 HLRWLVRSVTGQGDATGATG------------------------------
TH_0684|M.thermoresistible__bu HLRALAAAAADARRATS---------------------------------
MSMEG_2036|M.smegmatis_MC2_155 QLRKLAEAVISPQRLNS---------------------------------
Mb3196c|M.bovis_AF2122/97 HLRALAESVTRHVRDRRISS------------------------------
Rv3171c|M.tuberculosis_H37Rv HLRALAESVTRHVRDRRISS------------------------------
MMAR_1388|M.marinum_M HLRTLVDSVGGRSRAR-ISS------------------------------
MUL_2488|M.ulcerans_Agy99 HLRTLVDSVGGRSRAR-ISS------------------------------
MAV_2706|M.avium_104 EIEQLADATR----------------------------------------
MAB_3738c|M.abscessus_ATCC_199 GIDRLVRRSTPPLFAAIKQRLRDRTGL-----------------------
MLBr_00862|M.leprae_Br4923 AVHDLADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETAL
: *.
Mflv_4502|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1865|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0684|M.thermoresistible__bu --------------------------------------------------
MSMEG_2036|M.smegmatis_MC2_155 --------------------------------------------------
Mb3196c|M.bovis_AF2122/97 --------------------------------------------------
Rv3171c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1388|M.marinum_M --------------------------------------------------
MUL_2488|M.ulcerans_Agy99 --------------------------------------------------
MAV_2706|M.avium_104 --------------------------------------------------
MAB_3738c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 ALARRGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFA
Mflv_4502|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1865|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0684|M.thermoresistible__bu --------------------------------------------------
MSMEG_2036|M.smegmatis_MC2_155 --------------------------------------------------
Mb3196c|M.bovis_AF2122/97 --------------------------------------------------
Rv3171c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1388|M.marinum_M --------------------------------------------------
MUL_2488|M.ulcerans_Agy99 --------------------------------------------------
MAV_2706|M.avium_104 --------------------------------------------------
MAB_3738c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 EQVSAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRA
Mflv_4502|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1865|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0684|M.thermoresistible__bu --------------------------------------------------
MSMEG_2036|M.smegmatis_MC2_155 --------------------------------------------------
Mb3196c|M.bovis_AF2122/97 --------------------------------------------------
Rv3171c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1388|M.marinum_M --------------------------------------------------
MUL_2488|M.ulcerans_Agy99 --------------------------------------------------
MAV_2706|M.avium_104 --------------------------------------------------
MAB_3738c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 FGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAEL
Mflv_4502|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1865|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0684|M.thermoresistible__bu --------------------------------------------------
MSMEG_2036|M.smegmatis_MC2_155 --------------------------------------------------
Mb3196c|M.bovis_AF2122/97 --------------------------------------------------
Rv3171c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1388|M.marinum_M --------------------------------------------------
MUL_2488|M.ulcerans_Agy99 --------------------------------------------------
MAV_2706|M.avium_104 --------------------------------------------------
MAB_3738c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDRRGQLDMMFRLRR
Mflv_4502|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1865|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0684|M.thermoresistible__bu --------------------------------------------------
MSMEG_2036|M.smegmatis_MC2_155 --------------------------------------------------
Mb3196c|M.bovis_AF2122/97 --------------------------------------------------
Rv3171c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1388|M.marinum_M --------------------------------------------------
MUL_2488|M.ulcerans_Agy99 --------------------------------------------------
MAV_2706|M.avium_104 --------------------------------------------------
MAB_3738c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 YGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARIRVI