For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTATPEVDLPNPYTGTGTIHNPATGAVAGEVRWTDPADVPRIAAGLREAQKEWEARGPKGRAKVLARYAV WLGDHRDEIEKLLIAETGKSAVDAAQEVPLILMILSYYIKTVEKAMAPEKRPAPLPFLSIKKIEVHYRPR AVVGIIAPWNYPVANAMMDAIGALAAGCSVLLKPSERTPLTAEVLLRGWLDSGAPEVFALAQGARAVSEA VIDNSDYIQFTGSSATGAKVMERAARRLTPVSLELGGKDPMIVLEDADIELAANAAVWGAMFNAGQTCVS VERVYVLESVYDQFVAATVRAVENLKMGAGEGYDFGSLIDAGQVDITERHVNEALEKGAKALTGGKRPED GGSFYPPTVLVDVDHSMSCMNEETFGPTLPIMKVSSVEEAVRLANDSPYGLSASVFSKDTERAKKIALQL DCGAVNINDVISNLMCTTAPMGGWKTSGIGARFGGVDGVRKFCKQETVVAPRTSVGAGGTYYNNSHKALA RMNRLMTKLALARPRRTAK
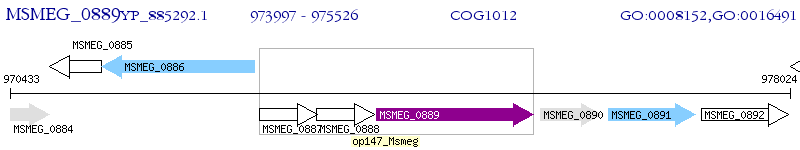
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0889 | - | - | 100% (509) | succinic semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5912 | gabD2 | 3e-84 | 38.69% (473) | succinic semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0309 | - | 1e-66 | 34.91% (464) | aldehyde dehydrogenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1760 | gabD2 | 2e-83 | 39.31% (463) | succinic semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0132 | - | 0.0 | 81.53% (509) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv1731 | gabD2 | 2e-83 | 39.31% (463) | succinic semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 4e-48 | 29.81% (463) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4136c | - | 1e-170 | 58.40% (500) | putative aldehyde dehydrogenase |
| M. marinum M | MMAR_1607 | putA_2 | 1e-117 | 47.84% (487) | succinate-semialdehyde dehydrogenase |
| M. avium 104 | MAV_4946 | - | 3e-68 | 36.21% (464) | aldehyde dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_3138 | - | 0.0 | 81.34% (509) | PUTATIVE aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_3430 | putA_2 | 1e-117 | 48.25% (487) | succinate-semialdehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0771 | - | 0.0 | 83.30% (509) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1760|M.bovis_AF2122/97 ---MPAPSAEVFDRLRNLAAIKDVAARPTRTIDEVFTGKPLTTIPVGTAA
Rv1731|M.tuberculosis_H37Rv ---MPAPSAEVFDRLRNLAAIKDVAARPTRTIDEVFTGKPLTTIPVGTAA
Mflv_0132|M.gilvum_PYR-GCK -------------MTATSDAHTANPFTGTGTITNPATGAVAGTVHWTDPA
Mvan_0771|M.vanbaalenii_PYR-1 ----------MGFMTATSDVQPPHTFTGTGTIRNPATGAVAGEVRWTDPA
MSMEG_0889|M.smegmatis_MC2_155 -------------MTATPEVDLPNPYTGTGTIHNPATGAVAGEVRWTDPA
TH_3138|M.thermoresistible__bu -------------MTATSGVDA----TETATIRNPATGGVAGEVRWTDPA
MAB_4136c|M.abscessus_ATCC_199 -------------MTTTSENLVSTPSDTSHDVRNPANGQVVGRVNWTDPS
MMAR_1607|M.marinum_M -----------MSSIATEPTTQKDDGQPQIPVENPATGEVIGHVPDMSAA
MUL_3430|M.ulcerans_Agy99 -----------MSSIATEPTMQKDDGQPQIPVENPATGEVIGHVPDMSAA
MAV_4946|M.avium_104 MSDIQTQAKTEYDKLFIGGKWTEPSTSEVIEVHCPATGEYVGKVPLAAAA
MLBr_02573|M.leprae_Br4923 ---------------------------MSIATINPATGETVKTFTPTTQD
.* .
Mb1760|M.bovis_AF2122/97 DVEAAFAEARAAQT--DWAKRPVIERAAVIRRYRDLVIENREFLMDLLQA
Rv1731|M.tuberculosis_H37Rv DVEAAFAEARAAQT--DWAKRPVIERAAVIRRYRDLVIENREFLMDLLQA
Mflv_0132|M.gilvum_PYR-GCK DVAQVAAGLRTAQR--EWEARGPKGRAKVLARYALWLGEHRAEIERLLIA
Mvan_0771|M.vanbaalenii_PYR-1 DVARLAAGLRTAQR--EWETRGPKGRAKVLARYAVWLGEHRDEIERLLIA
MSMEG_0889|M.smegmatis_MC2_155 DVPRIAAGLREAQK--EWEARGPKGRAKVLARYAVWLGDHRDEIEKLLIA
TH_3138|M.thermoresistible__bu DVPRITAGLREAQR--HWEARGPKGRAKVLARYAVWLGEHRDEIERLLIA
MAB_4136c|M.abscessus_ATCC_199 DIPGIVAQLAKAQP--SWERLGPEGRGQVLARFAQWLIDNTGRIEAQLIA
MMAR_1607|M.marinum_M QIAELARRGRAAQP--GWEALGYEGRAVVLKRMQRWLVDNADQVIATICS
MUL_3430|M.ulcerans_Agy99 QIAELARRGRAAQP--GWEALGYEGRAVVLKRMQRWLVDNADQVIATICS
MAV_4946|M.avium_104 DVDAAVAAARAAFDNGPWPTTPPKERAAVIANALKVMEERKDLFTGLLAA
MLBr_02573|M.leprae_Br4923 EVEDAIARAYARFD--NYRHTTFTQRAKWANATADLLEAEANQSAALMTL
:: : *. : . :
Mb1760|M.bovis_AF2122/97 EAGKARWAAQEEIVDL-IANANYYARVCVDLLKP--RKAQPLLPGIGKTT
Rv1731|M.tuberculosis_H37Rv EAGKARWAAQEEIVDL-IANANYYARVCVDLLKP--RKAQPLLPGIGKTT
Mflv_0132|M.gilvum_PYR-GCK ETGKSAVDAAQEVPLI-LMITSYYVRTMEKALAPESRPAALPFLSIKKIT
Mvan_0771|M.vanbaalenii_PYR-1 ETGKSAADAAQEVPLI-IMITSYYVRTMAKALAPETRPASLPFLSIKKIT
MSMEG_0889|M.smegmatis_MC2_155 ETGKSAVDAAQEVPLI-LMILSYYIKTVEKAMAPEKRPAPLPFLSIKKIE
TH_3138|M.thermoresistible__bu ETGKSAVDAAQEVPLI-LMITSYYIKTMEKALAPETRPASMPLLKIKKIT
MAB_4136c|M.abscessus_ATCC_199 ETGKSKIDGGIEIPSI-LAIIAYYAPKAAEFLAPESRPASSIAMNTKKIT
MMAR_1607|M.marinum_M ETGKTYEDAQIAELIYGAAAFGFWADKAPRYLADERIHSSSVFVKGKRLV
MUL_3430|M.ulcerans_Agy99 ETGKTYEDAQIAELIYGAAAFGFWADKAPRYLADERIHSSSVFVKGKRLV
MAV_4946|M.avium_104 ETGQPPTSVETMHWMSSIGALNFFAGPAVDQVKWKEVRTG----GYGQSI
MLBr_02573|M.leprae_Br4923 EMGKTLQSAKDEAMKC-AKGFRYYAEKAETLLAD--EPAEAAAVGASKAL
* *:. :: : : :
Mb1760|M.bovis_AF2122/97 VCYQPKGVVGVISPWNYPMTLTVSDSVPALVAGNAVVLKPDSQTPYCALA
Rv1731|M.tuberculosis_H37Rv VCYQPKGVVGVISPWNYPMTLTVSDSVPALVAGNAVVLKPDSQTPYCALA
Mflv_0132|M.gilvum_PYR-GCK VHYRPRPVVGIIAPWNYPVANALMDAIGALAAGCAVLLKPSERTPLTAEL
Mvan_0771|M.vanbaalenii_PYR-1 VHYRPRPVVGIIAPWNYPVANALMDAVGALAAGCAVLLKPSERTPLTAEL
MSMEG_0889|M.smegmatis_MC2_155 VHYRPRAVVGIIAPWNYPVANAMMDAIGALAAGCSVLLKPSERTPLTAEV
TH_3138|M.thermoresistible__bu VHHRPRPVVGVIAPWNYPVANALMDAIGALAAGCAVLLKPSERTPLTAEL
MAB_4136c|M.abscessus_ATCC_199 VHQRPRKVVGVISPWNYPVALGYWDVIPALLAGCSVLLKPSERTPLSDEL
MMAR_1607|M.marinum_M LRYRPMGLIGVIGPWNYPLTNSFGDCIPALAAGNSVILKPSEVTPLTSLL
MUL_3430|M.ulcerans_Agy99 LRYRPMGLIGVIGPWNYPLTNSFGDCIPALAAGNSVILKPSEVTPLTSLL
MAV_4946|M.avium_104 VHREPIGVVGAIVAWNVPLFLAVNKLGPALLAGCTVVLKPAAETPLSANA
MLBr_02573|M.leprae_Br4923 ARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQCALY
.* :: : .** *: ** ** :** .*
Mb1760|M.bovis_AF2122/97 CAELLYRAGLPRALYAIVP-GPGSVVGTAITDNCDYLMFTGSSATGSRLA
Rv1731|M.tuberculosis_H37Rv CAELLYRAGLPRALYAIVP-GPGSVVGTAITDNCDYLMFTGSSATGSRLA
Mflv_0132|M.gilvum_PYR-GCK LLRGWLEAGGPD-VLALAQ-GAR-AVSEAVIDNSDFIQFTGSSATGAKVM
Mvan_0771|M.vanbaalenii_PYR-1 LLRGWLDSGGPD-VLALAQ-GAR-EVSEAVIDNSDYIQFTGSTATGVKVM
MSMEG_0889|M.smegmatis_MC2_155 LLRGWLDSGAPE-VFALAQ-GAR-AVSEAVIDNSDYIQFTGSSATGAKVM
TH_3138|M.thermoresistible__bu LLRGWLESGAPE-VFALAQ-GAR-EVSEAVVDNADFIQFTGSTATGIKVM
MAB_4136c|M.abscessus_ATCC_199 IANGWAEIGGPP-VFEVVH-GAR-EVGEALVDNVDFVQFTGSTATGKKIM
MMAR_1607|M.marinum_M LAQGLAESGLPDGVFAVAT-GRG-GTGAAVIDEVDMIMFTGSTATGRKVA
MUL_3430|M.ulcerans_Agy99 LAQGLAESGLPDGVFAVAT-GRG-GTGAAVIDEVDMIMFTGSTATGRKVA
MAV_4946|M.avium_104 LAEAFAEAGLPEGVLSVVPGGIETGQALTSNTDVDIFSFTGSSAVGREIG
MLBr_02573|M.leprae_Br4923 LADVIARGGFPEDCFQTLLISANGVEAILRDPRVAAATLTGSEPAGQAVG
* * . . :*** ..* :
Mb1760|M.bovis_AF2122/97 EHAGRRLIGFSAELGGKNPMIVARGANLDKVAKAATRACFSNAGQLCISI
Rv1731|M.tuberculosis_H37Rv EHAGRRLIGFSAELGGKNPMIVARGANLDKVAKAATRACFSNAGQLCISI
Mflv_0132|M.gilvum_PYR-GCK ERAARRLTPVSLELGGKDPMIVLEDADVDLAAHAAVWGAMFNAGQTCVSV
Mvan_0771|M.vanbaalenii_PYR-1 ERAARRLTPVSLELGGKDPMIVLEDADIDLAAHAAVWGAMFNAGQTCVSV
MSMEG_0889|M.smegmatis_MC2_155 ERAARRLTPVSLELGGKDPMIVLEDADIELAANAAVWGAMFNAGQTCVSV
TH_3138|M.thermoresistible__bu ERAARRLTPVSLELGGKDPMIVLEDADIELAANAAVWGAMFNAGQTCVSV
MAB_4136c|M.abscessus_ATCC_199 ERAARRLTPVSLELGGKDPMLVLPDADVKRAAHAAVWGSMFNAGQTCVSV
MMAR_1607|M.marinum_M VRAAERLIPASLELGGKDPMIVLADADIERAANHAAYYSMFNCGQTCISI
MUL_3430|M.ulcerans_Agy99 VRAAERLIPASLELGGKDPMIVLADADIERAANHAAYYSMFNCGQTCISI
MAV_4946|M.avium_104 RRAADMLKPCTLELGGKSAAIVLDDVDLPSALPMLVFSGIMNSGQACVAQ
MLBr_02573|M.leprae_Br4923 AIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRVQNNGQSCIAA
*. : ****... :* ..:: . . . * ** *::
Mb1760|M.bovis_AF2122/97 ERIYVEKDIAEEFTRKFGDAVRNMKLGTAYDFS-VDMGSLISEAQLKTVS
Rv1731|M.tuberculosis_H37Rv ERIYVEKDIAEEFTRKFGDAVRNMKLGTAYDFS-VDMGSLISEAQLKTVS
Mflv_0132|M.gilvum_PYR-GCK ERVYVLDQVYDQFVDAVVRDVAKLQVGAGEGN---AFGALIDDDQVAVTE
Mvan_0771|M.vanbaalenii_PYR-1 ERVYVLEQVYDQFVAAVVRDVESLKMGAGEGN---AFGALIDDSQVEITA
MSMEG_0889|M.smegmatis_MC2_155 ERVYVLESVYDQFVAATVRAVENLKMGAGEGY---DFGSLIDAGQVDITE
TH_3138|M.thermoresistible__bu ERVYVLAPVYEQFVAAVVRAVEKLEVGAGEGR---HFGALIDESQLAVTE
MAB_4136c|M.abscessus_ATCC_199 ERVYVHDSIYEQFVDLVVEEVRGLEIGAGLDH---SVGAMIDENQLEVVD
MMAR_1607|M.marinum_M ERCYVEAPVYDEFVAKVTEKVRAIRQGVPGGPGSVDVGSLTFPPQVETVQ
MUL_3430|M.ulcerans_Agy99 ERCYVEAPVYDEFVAKVTEKVRAIRQGVPAGPGSVDVGSLTFPPQVETVQ
MAV_4946|M.avium_104 TRILAPRSRYDEIVDAVSTFVQSLPVGLPSDP-AAQIGSLISEKQRARVE
MLBr_02573|M.leprae_Br4923 KRFIAHADIYDEFVEKFVARMETLRVGDPTDP-DTDVGPLATESGRAQVE
* . :::. : : * . .*.: .
Mb1760|M.bovis_AF2122/97 GHVDDATAKGAKVIAGGKARPDIGP-LFYEPTVLTNVAPEMECAANETFG
Rv1731|M.tuberculosis_H37Rv GHVDDATAKGAKVIAGGKARPDIGP-LFYEPTVLTNVAPEMECAANETFG
Mflv_0132|M.gilvum_PYR-GCK RHVADALAKGARALTGGTRGAGSG--SFYEPTVLVDVDHAMLCMTEETFG
Mvan_0771|M.vanbaalenii_PYR-1 RHVEDALAKGARALTGGKRGSGPG--SFYEPTVLVDVDHSMACMTEETFG
MSMEG_0889|M.smegmatis_MC2_155 RHVNEALEKGAKALTGGKRPEDGG--SFYPPTVLVDVDHSMSCMNEETFG
TH_3138|M.thermoresistible__bu RHVADAVARGAKVLTGGRRKDGPG--CFYEPTVLVDVDHSMMCMTEETFG
MAB_4136c|M.abscessus_ATCC_199 RHVRQAVSSGARALTGGARIPGHG--SFYAPTVLVDVDHSMDCMREETFG
MMAR_1607|M.marinum_M RHVEEAVERGAKVLTGGHRGRTKG--YWYEPTVLVDVNHDMACMREETFG
MUL_3430|M.ulcerans_Agy99 RHVEEAVERGAKVLTGGHRGRTKG--YWYEPTVLVDVNHDMACMREETFG
MAV_4946|M.avium_104 GYIAKGIEEGARLVCGGGRPEGLDSGFFVQPTVFADVDNKMTIAQEEIFG
MLBr_02573|M.leprae_Br4923 QQVDAAAAAGAVIRCGGKRPDRPG--WFYPPTVITDITKDMALYTDEVFG
: . ** ** . : ***:.:: * :* **
Mb1760|M.bovis_AF2122/97 PVVSIYPVADVDEAVEKANDTDYGLNASVWAGSTAEGQRIAARLRSGTVN
Rv1731|M.tuberculosis_H37Rv PVVSIYPVADVDEAVEKANDTDYGLNASVWAGSTAEGQRIAARLRSGTVN
Mflv_0132|M.gilvum_PYR-GCK PTLPIMRVSTVDEAVRLANDSPYGLSAAVFSKDLERAEAVAVQLDCGGVN
Mvan_0771|M.vanbaalenii_PYR-1 PTLPIMKVSSVEEAVRLANDSPYGLSAAVFSRDVDRAKAVALQLDCGGVN
MSMEG_0889|M.smegmatis_MC2_155 PTLPIMKVSSVEEAVRLANDSPYGLSASVFSKDTERAKKIALQLDCGAVN
TH_3138|M.thermoresistible__bu PTLPIMKVSSVAEAVRLANDTPYGLSGSVFSQDVERAREIALQLDCGAVN
MAB_4136c|M.abscessus_ATCC_199 PTLPIMRYSEESEAIALANDSEYGLSASVWSKDRRRAERVALQLDCGTVN
MMAR_1607|M.marinum_M PTLPIMKVADAEEAIRLANDSEYGLCAAVFSRDLAHAEAVARRVNSGAVT
MUL_3430|M.ulcerans_Agy99 PTLPIMKVADAEEAIRLANDSEYGLCAAVFSRDLAHAEAVARRVNSGAVT
MAV_4946|M.avium_104 PVLSIIPYDTEEDAIKIANDSVYGLAGSVWTADVQRGIAVSEKIRTGTYA
MLBr_02573|M.leprae_Br4923 PVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNIVAGQVF
*. .: :*: ** : :** . .:: . . . .: *
Mb1760|M.bovis_AF2122/97 VDEGYAFAWGSLSAPMGGMG-LSGVGRRHG-PEGLLKYTESQTIATARVF
Rv1731|M.tuberculosis_H37Rv VDEGYAFAWGSLSAPMGGMG-LSGVGRRHG-PEGLLKYTESQTIATARVF
Mflv_0132|M.gilvum_PYR-GCK IND-VISNLMCTTAPMGGWK-TSGIGARFGGPEGLRKYCRIETVVAPRTN
Mvan_0771|M.vanbaalenii_PYR-1 VND-VISNLMCTTAPMGGWK-SSGIGARFGGPEGLRKYCRIETVVSPRTN
MSMEG_0889|M.smegmatis_MC2_155 IND-VISNLMCTTAPMGGWK-TSGIGARFGGVDGVRKFCKQETVVAPRTS
TH_3138|M.thermoresistible__bu IND-VISNLMATTAPMGGWK-TSGIGARFGGPEGLRKYCRVETVVEPRTG
MAB_4136c|M.abscessus_ATCC_199 IND-VIMNLSCLTAPHGGWK-GSGLGSRFGGVDGLRKYCRTEAIVDTRVT
MMAR_1607|M.marinum_M VND-ALLNYTALELPMGGAKPGSGVGYRHG-PGGIRKYCRQQALLISRFH
MUL_3430|M.ulcerans_Agy99 VND-ALLNYTALELPMGGAKPGSGVGYRHG-PGGIRKYCRQQALLISRFH
MAV_4946|M.avium_104 INW----YAFDPCCPFGGYK-NSGIGRENG-PEGVEHFTQQKSVLMPMGY
MLBr_02573|M.leprae_Br4923 ING---MTVSYPELPFGGIK-RSGYGRELS-THGIREFCNIKTVWIA---
:: * ** ** * . . *: .: . ::: .
Mb1760|M.bovis_AF2122/97 NLDPPFGIPATVWQKSLLPIVRTVMKLPGRR----
Rv1731|M.tuberculosis_H37Rv NLDPPFGIPATVWQKSLLPIVRTVMKLPGRR----
Mflv_0132|M.gilvum_PYR-GCK VGAGGNYYNNSPRALKRMNTMMTKLALIRPRRVAK
Mvan_0771|M.vanbaalenii_PYR-1 VGAGGNYYNNSLRALKRMNKLMTKLALIRPRRVAK
MSMEG_0889|M.smegmatis_MC2_155 VGAGGTYYNNSHKALARMNRLMTKLALARPRRTAK
TH_3138|M.thermoresistible__bu IGAGGTYYNNSFKSLGMMNKLLTRMALVRPRRLAK
MAB_4136c|M.abscessus_ATCC_199 LPSEPLWYSGPSWLAPVALKSLTKLSNKALRPFRR
MMAR_1607|M.marinum_M ARRDVHMYPYSARRTRILGRLVGRLYRG-VRGTDS
MUL_3430|M.ulcerans_Agy99 ARRDVHMYPYSARRTRILGRLVGRLYRG-VRGTDS
MAV_4946|M.avium_104 TLEG-------------------------------
MLBr_02573|M.leprae_Br4923 -----------------------------------