For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTAISTTATDLDVALGDPFCPANPHGATAILAADERAERALATYQLLADLGLQRMFVPIELGGDLVDIPS LVRVLAPVFGRDMGLGLGFGVTTLMAALNVWHSGDDRQRRRVADQVLTGAPLSVAYHELDHGNDMAANEV RAVATAGGYRLSGRKEVINNASRASAAVVFARTSDRPGRSHSLFLADLDNVRRLDRFRTTALRTAQLGGL DFHDHQVGVEDRIGAEGHGIEIALKSFQISRVVMAGAGLGPVDVALHLAASFARQRVVYGRRVDQIPHAR TNLAIAQSLLLAVEAAVTTAAVGLHTSPQHAGAQAASVKYAAPLLLEYAMEHLAVVLGARFYLRTGPFAL FGKHYRDLAPVGIGHAGSTSCLLALLPQVRHLLQPGTAAALPAPGAQLPRLDFSELVLRSGAPDPIVSWT PAATPLTAPGAGDLIEEQARTLERLRGAAERIPASALGVTAPPAAFDLAEQFTKRFVVAAALAVRESDRT EFADAWVRIAIAAVAAHRRGRAVVDPADVDVVVQRIDRHVAGSRSLLLDGHELPRSIPAL*
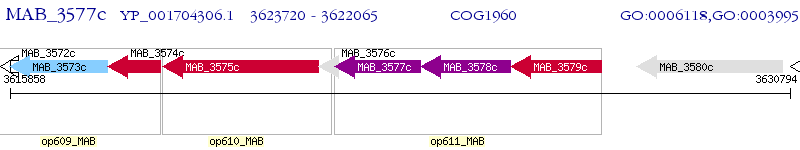
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3577c | - | - | 100% (551) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3578c | - | 1e-40 | 30.37% (540) | acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2085 | - | 2e-15 | 25.93% (324) | acyl CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2743c | fadE20 | 3e-15 | 28.42% (278) | acyl-CoA dehydrogenase FADE20 |
| M. gilvum PYR-GCK | Mflv_4943 | - | 1e-15 | 26.74% (258) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv2724c | fadE20 | 2e-15 | 28.42% (278) | acyl-CoA dehydrogenase FADE20 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 2e-09 | 22.77% (325) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_1989 | fadE20 | 4e-14 | 27.34% (278) | acyl-CoA dehydrogenase FadE20 |
| M. avium 104 | MAV_2703 | - | 3e-18 | 26.95% (308) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4832 | - | 3e-13 | 25.36% (343) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2583 | - | 1e-15 | 26.17% (298) | PUTATIVE peptidase S1 and S6, chymotrypsin/Hap |
| M. ulcerans Agy99 | MUL_3367 | fadE20 | 2e-14 | 26.98% (278) | acyl-CoA dehydrogenase FadE20 |
| M. vanbaalenii PYR-1 | Mvan_1476 | - | 2e-16 | 27.52% (258) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1989|M.marinum_M -----MGSAIKYQRTLFEPEHDLFRESYRGFLERHVAPYHDEWEKAKIVD
MUL_3367|M.ulcerans_Agy99 -----MGSAIKYQRTLFEPEHDLFRESYRGFLERHVALYHDEWEKAKIVD
Mb2743c|M.bovis_AF2122/97 -----MGSATKYQRTLFEPEHELFRESYRAFLDRHVAAYHDEWEKTKIVD
Rv2724c|M.tuberculosis_H37Rv -----MGSATKYQRTLFEPEHELFRESYRAFLDRHVAPYHDEWEKTKIVD
TH_2583|M.thermoresistible__bu MSGIDTGRAVFYDRTLFEPEHEMFRDSYRTFLKRSVAPYHQQWEIDGIVD
MSMEG_4832|M.smegmatis_MC2_155 -----------MQRNLFTEDHEAFRTLARDFIEKEVVSAYPEWEKAGRMP
MAV_2703|M.avium_104 ----MSELFPDYRPSWETDQHRELRKHAAEFFRKEATPNQERWAAQHAVD
Mflv_4943|M.gilvum_PYR-GCK --MP-------VDRLLPTDDARDLVELARQIADKVLDPIVDRHEKDETYP
Mvan_1476|M.vanbaalenii_PYR-1 --MA-------VDRLLPTDEARELVDLARQIADKALDPIVDQHEKDETYP
MLBr_00737|M.leprae_Br4923 --MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFP
MAB_3577c|M.abscessus_ATCC_199 ---------MTTAISTTATDLDVALGDPFCPANPHGATAILAADERAERA
:
MMAR_1989|M.marinum_M RGVWLEAGKQGFLGMAVPEEYGGGGNPDFRYNTIITEETSAGRYSGIGFG
MUL_3367|M.ulcerans_Agy99 RGVWLEAGKQGFLGMAVPEEYGGGGNPDFRYNTIITEETSAGRYSGIGFG
Mb2743c|M.bovis_AF2122/97 RGVWLEAGKQGFLGMAVPEEYGGGGNADFRYNTVITEETCAGRYSGIGFG
Rv2724c|M.tuberculosis_H37Rv RGVWLEAGKQGFLGMAVPEEYGGGGNADFRYNTVITEETCAGRYSGIGFG
TH_2583|M.thermoresistible__bu RGVWLEAGKQGFLGMGVPEEHGGGGVADFRYNAIVTEETTAAGFSGLGFA
MSMEG_4832|M.smegmatis_MC2_155 RDVFKQMGELGMLGMAIPEEYGGAGIPDYRYNVVLQEEAARALVTLSTVR
MAV_2703|M.avium_104 REFWNKAGDAGLLGLDLPEEYGGAG-GDFAMNVVVQEELAYAYDQGFGFA
Mflv_4943|M.gilvum_PYR-GCK EGVFATLGEAGLLSLPYPEEWGGGGQPYEVYLQVLEELAARWAAVAVAVS
Mvan_1476|M.vanbaalenii_PYR-1 DGVFATLGEAGLLSLPYPEEWGGGGQPYEVYLQVLEELAARWAAVAVAVS
MLBr_00737|M.leprae_Br4923 EEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPA
MAB_3577c|M.abscessus_ATCC_199 LATYQLLADLGLQRMFVPIELGGDLVDIPSLVRVLAPVFGRDMGLGLGFG
*: : * * ** ::
MMAR_1989|M.marinum_M LHNDIVAPYLLALATEEQKQRWLPKFCTGEFITAIAMTEPGTGSDLQGIK
MUL_3367|M.ulcerans_Agy99 LHNEIVAPYLLALATEEQKQRRLPKFCTGEFITAIAMTEPGTGSDLQGIK
Mb2743c|M.bovis_AF2122/97 LHNDIVAPYLLALATEEQKRRWFPNFCTGELTTAIAMTEPGTGSDLQGIT
Rv2724c|M.tuberculosis_H37Rv LHNDIVAPYLLALATEEQKRRWFPNFCTGELITAIAMTEPGTGSDLQGIT
TH_2583|M.thermoresistible__bu LHNDMVAPYLLRLATAEQKQRWLPRFCSGECITALAMTEPGAGSDLQGIR
MSMEG_4832|M.smegmatis_MC2_155 TQLEVILPYFLHYADTEQRQRWFPGLAAGTLLTAIAMTEPGTGSDLAGVR
MAV_2703|M.avium_104 VHSPIVAHYIWAYGSDEQRRRWLPKIISGEAVLAIAMTEPGTGSDLQSVK
Mflv_4943|M.gilvum_PYR-GCK VH-SLSCHPLMSFGSDEQRQRWLPDMLGGSTIGAYSLSEPQAGSDAAALT
Mvan_1476|M.vanbaalenii_PYR-1 VH-SLSCHPLMSFGTDEQRQRWLPEMLGGSTIGAYSLSEPQAGSDAAALT
MLBr_00737|M.leprae_Br4923 VN-KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMR
MAB_3577c|M.abscessus_ATCC_199 VTTLMAALNVWHSGDDRQRRRVADQVLTGAPLS-VAYHELDHGNDMAANE
: . . . ::: . : * *.* .
MMAR_1989|M.marinum_M TRAVKQGDHYLLNGSKTFITNGINADLVIVVAQT---DPEKGAQGFSLLV
MUL_3367|M.ulcerans_Agy99 TRAVKQGDHYLLNGSKTFITNGINADLVIVVAQT---DPEKGAQGFSLLV
Mb2743c|M.bovis_AF2122/97 TRAVKHGDHYVLNGSKTFITNGINSDLVIVVAQT---DPEKGAQGFSLLV
Rv2724c|M.tuberculosis_H37Rv TRAVKHGDHYVLNGSKTFITNGINSDLVIVVAQT---DPEKGAQGFSLLV
TH_2583|M.thermoresistible__bu TRAVRDGDGWVLNGSKTFITNGINADLVIVFAQT---DPDKGSMGYSLLV
MSMEG_4832|M.smegmatis_MC2_155 TTAVRDGDHYVVNGAKTFITGGIQADLVVVVARTS-TDPDNRRKGLTLLV
MAV_2703|M.avium_104 TTARRDGDFYVINGSKTFISNGTHCDLLVIVAKT---DPSKGAAGVSLIV
Mflv_4943|M.gilvum_PYR-GCK CRATAVDGGYRITGSKAWITHGGIADFYNLFARTG-----EGSQGISCFL
Mvan_1476|M.vanbaalenii_PYR-1 CKAVAADGGYRINGAKAWITHGGIADFYNLFARTG-----EGSQGISCFL
MLBr_00737|M.leprae_Br4923 TRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDP---DKGANGISAFI
MAB_3577c|M.abscessus_ATCC_199 VRAVATAGGYRLSGRKEVINNASRASAAVVFARTS----DRPGRSHSLFL
* . : :.* * *. . . :.* * . . : ::
MMAR_1989|M.marinum_M VER--GMEGFERGRHLDKIGLDAQDTAELSFSDVQVPVENLLG-QEGMGF
MUL_3367|M.ulcerans_Agy99 VER--GMEGFERGRHLDKIGLDAQDTAELSFSDVQVPVENLLG-QEGMGF
Mb2743c|M.bovis_AF2122/97 VER--GMAGFERGRQLDKIGLDAQDTAELSFTDVAVPAENLLG-QEGMGF
Rv2724c|M.tuberculosis_H37Rv VER--GMAGFERGRQLDKIGLDAQDTAELSFTDVAVPAENLLG-QEGMGF
TH_2583|M.thermoresistible__bu VER--GMAGFERGRRLDKIGMTGQDTAELSFTDVRVPAENLLG-KVGKGL
MSMEG_4832|M.smegmatis_MC2_155 VED--GMAGFSRGRELEKMGCKVQDTAELSFTDVRVPVANVLG-EEGEAF
MAV_2703|M.avium_104 AETDENLAGFERGRVLAKMGQHAQDTRELFFSDMRVPVANLLGAQEGLGF
Mflv_4943|M.gilvum_PYR-GCK VPR--DTEGLTFGKPEEKMGLHAVPTTAAHYDDANLDSDRRIG-TEGQGL
Mvan_1476|M.vanbaalenii_PYR-1 VPR--DTEGLTFGKPEEKMGLHAVPTTAAHYENAFVEADRRIG-AEGQGL
MLBr_00737|M.leprae_Br4923 VHK--DDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIG-EPGTGF
MAB_3577c|M.abscessus_ATCC_199 ADL----DNVRRLDRFRTTALRTAQLGGLDFHDHQVGVEDRIG-AEGHGI
. .. . . : . : :* * .:
MMAR_1989|M.marinum_M IYLMQNLPQERISIAIMAAAGMESVLDQTLQYTKERKAFGRSIGNFQNSR
MUL_3367|M.ulcerans_Agy99 IYLMQNLPQERISIAIMAAAGMESVLDQTLQYTKERKAFGRSIGNFQNSR
Mb2743c|M.bovis_AF2122/97 IYLMQNLPQERISIAIMAAAGMESVLEQTLQYAKERKAFGRSIGSFQNSR
Rv2724c|M.tuberculosis_H37Rv IYLMQNLPQERISIAIMAAAGMESVLEQTLQYAKERKAFGRSIGSFQNSR
TH_2583|M.thermoresistible__bu LYLMQNLPQERLSIAVMSAAAMAATLESTLEYVQQRKAFGRPIGSFQNSR
MSMEG_4832|M.smegmatis_MC2_155 SYLGHNLAQERLTVAVGSVAQARSAIDAAIDYTKTRKAFGTPVASFQNTK
MAV_2703|M.avium_104 YQLMEQLARERLIIATICAALAEVAVLEAIKYSREREAFGRPIGKFQHTK
Mflv_4943|M.gilvum_PYR-GCK QIAFSALDSGRLGIAAVAVGLAQAALDEAVAYSQERTTFGRKIIDHQGLA
Mvan_1476|M.vanbaalenii_PYR-1 QIAFSALDSGRLGIAAVAVGLAQAALDAAVDYSQERTTFGRKIIDHQGLA
MLBr_00737|M.leprae_Br4923 KTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQ
MAB_3577c|M.abscessus_ATCC_199 EIALKSFQISRVVMAGAGLGPVDVALHLAASFARQRVVYGRRVDQIPHAR
: * :. . .: : : : * :* :
MMAR_1989|M.marinum_M FVLAELATEATMVRIMVDEFIELHLDGKL-TAEQAAMAKWYSTEKQVSLN
MUL_3367|M.ulcerans_Agy99 FVLAELATEATMVRIMVDESIELHLDGKL-TAEQAAMAKWYSTEKQVSLN
Mb2743c|M.bovis_AF2122/97 FLLAELATEATVVRIMVDEFIKLHLAGKL-TAEQAAMAKWYATEKQVYLN
Rv2724c|M.tuberculosis_H37Rv FLLAELATEATVVRIMVDEFIKLHLAGKL-TAEQAAMAKWYATEKQVYLN
TH_2583|M.thermoresistible__bu FVLAELATEATAIRIMVDEFVRLHDAGKL-TAEQAAMAKWFATEAFVRLA
MSMEG_4832|M.smegmatis_MC2_155 FELAACSAEVEAAQAMLDRAVALHVDGEL-SGADAARVKLFCTEMQARVV
MAV_2703|M.avium_104 FVLAECKTEALAIKTMVDYAVGQYLDGKN-DPATASMAKLFAAEKSDQIV
Mflv_4943|M.gilvum_PYR-GCK FLLADMAAAVDSARATYLDAARRRDAGLP-YSRHASVAKLVATDAAMKVT
Mvan_1476|M.vanbaalenii_PYR-1 FLLADMAAAVDAARATYLDAARRRDAGLP-YSRHASVAKLTATDAAMKVT
MLBr_00737|M.leprae_Br4923 FMLADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVT
MAB_3577c|M.abscessus_ATCC_199 TNLAIAQSLLLAVEAAVTTAAVGLHTSPQHAGAQAASVKYAAPLLLEYAM
** . . :: * ..
MMAR_1989|M.marinum_M DRCLQLHGGYGYMREYPVARAYLDSR----VQTIYGGTTEIMKEIIGRSL
MUL_3367|M.ulcerans_Agy99 DRCLQLHGGYGYMREYPVARAYLDSR----VQTIYGGTTEIMKEIIGRSL
Mb2743c|M.bovis_AF2122/97 DRCLQLHGGYGYMREYPVARAYLDSR----VQTIYGGTTEIMKEIIGRGL
Rv2724c|M.tuberculosis_H37Rv DRCLQLHGGYGYMREYPVARAYLDSR----VQTIYGGTTEIMKEIIGRGL
TH_2583|M.thermoresistible__bu DRCLQLHGGYGYMREYPIAKSYLDAR----VQTIYGGTTEIMKEIIGRRL
MSMEG_4832|M.smegmatis_MC2_155 DRCLQLFGGYGYMMEYPIARLYTDAR----VARIYAGTSEVMKVIIAKSL
MAV_2703|M.avium_104 DKCLQIFGGYGYMTEYPIANMYTASR----VNRKYGRTSEIMKEIISRSL
Mflv_4943|M.gilvum_PYR-GCK TDAVQVLGGAGYTRDYRVERYMREAK----ITQIFEGTNQIQRLVISRHL
Mvan_1476|M.vanbaalenii_PYR-1 TDAVQVFGGAGYTRDYRVERYMREAK----ITQIFEGTNQIQRLVISRHL
MLBr_00737|M.leprae_Br4923 TDAVQLFGGAGYTSDFPVERFMRDAK----ITQIYEGTNQIQRVVMSRAL
MAB_3577c|M.abscessus_ATCC_199 EHLAVVLGARFYLRTGPFALFGKHYRDLAPVGIGHAGSTSCLLALLPQVR
: *. * . : : . :.. :: :
MMAR_1989|M.marinum_M GI------------------------------------------------
MUL_3367|M.ulcerans_Agy99 GI------------------------------------------------
Mb2743c|M.bovis_AF2122/97 GV------------------------------------------------
Rv2724c|M.tuberculosis_H37Rv GV------------------------------------------------
TH_2583|M.thermoresistible__bu CT------------------------------------------------
MSMEG_4832|M.smegmatis_MC2_155 GL------------------------------------------------
MAV_2703|M.avium_104 --------------------------------------------------
Mflv_4943|M.gilvum_PYR-GCK TRG-----------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 ARD-----------------------------------------------
MLBr_00737|M.leprae_Br4923 LR------------------------------------------------
MAB_3577c|M.abscessus_ATCC_199 HLLQPGTAAALPAPGAQLPRLDFSELVLRSGAPDPIVSWTPAATPLTAPG
MMAR_1989|M.marinum_M --------------------------------------------------
MUL_3367|M.ulcerans_Agy99 --------------------------------------------------
Mb2743c|M.bovis_AF2122/97 --------------------------------------------------
Rv2724c|M.tuberculosis_H37Rv --------------------------------------------------
TH_2583|M.thermoresistible__bu --------------------------------------------------
MSMEG_4832|M.smegmatis_MC2_155 --------------------------------------------------
MAV_2703|M.avium_104 --------------------------------------------------
Mflv_4943|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_3577c|M.abscessus_ATCC_199 AGDLIEEQARTLERLRGAAERIPASALGVTAPPAAFDLAEQFTKRFVVAA
MMAR_1989|M.marinum_M --------------------------------------------------
MUL_3367|M.ulcerans_Agy99 --------------------------------------------------
Mb2743c|M.bovis_AF2122/97 --------------------------------------------------
Rv2724c|M.tuberculosis_H37Rv --------------------------------------------------
TH_2583|M.thermoresistible__bu --------------------------------------------------
MSMEG_4832|M.smegmatis_MC2_155 --------------------------------------------------
MAV_2703|M.avium_104 --------------------------------------------------
Mflv_4943|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MAB_3577c|M.abscessus_ATCC_199 ALAVRESDRTEFADAWVRIAIAAVAAHRRGRAVVDPADVDVVVQRIDRHV
MMAR_1989|M.marinum_M --------------------
MUL_3367|M.ulcerans_Agy99 --------------------
Mb2743c|M.bovis_AF2122/97 --------------------
Rv2724c|M.tuberculosis_H37Rv --------------------
TH_2583|M.thermoresistible__bu --------------------
MSMEG_4832|M.smegmatis_MC2_155 --------------------
MAV_2703|M.avium_104 --------------------
Mflv_4943|M.gilvum_PYR-GCK --------------------
Mvan_1476|M.vanbaalenii_PYR-1 --------------------
MLBr_00737|M.leprae_Br4923 --------------------
MAB_3577c|M.abscessus_ATCC_199 AGSRSLLLDGHELPRSIPAL