For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPIATINPATGETIKTFTAASDAEIDAAIGRAHDRFRQYRTTSFSDRAAWALSTADLLEAEADDVAALMT LEMGKTLASAKAEVLKCAKGFRFYAENAEAMLAPEPADASAVNASQAYAQYQPLGVVLAVMPWNFPLWQA VRFAAPALMAGNVGLLKHASNVPQSALYLADVIARGGFPEGCFQTLLVPSSAVERILRDPRVAAATLTGS EPAGRSVAAIAGDEVKPTVLELGGSDPFIVMPSADLEQAVSTAVTARVQNNGQSCIAAKRFIAHRDIYDD FLSRFVEKMAALRVGDPTDPATDVGPLATEQGRDEVDKQVQQAVAAGASIRCGGTRPDGPGWYYPPTVIT DITKNMPIFGEEVFGPVASVYRATDIEEAIEVANATSFGLGANAWTRETAEQQRFINELDAGQVFINGMT VSYPEVPFGGIKRSGYGRELSAHGIREFCNIKTVWIA
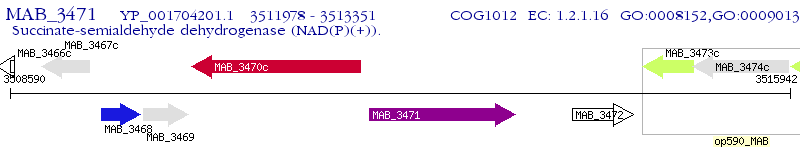
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3471 | gabD1 | - | 100% (457) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4322 | - | 3e-98 | 42.26% (452) | aldehyde dehydrogenase family protein |
| M. abscessus ATCC 19977 | MAB_4418 | - | 2e-77 | 36.82% (459) | succinate-semialdehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0239c | gabD1 | 0.0 | 81.62% (457) | succinic semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4860 | - | 2e-73 | 35.48% (451) | gamma-aminobutyraldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0234c | gabD1 | 0.0 | 81.62% (457) | succinic semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 0.0 | 77.02% (457) | succinic semialdehyde dehydrogenase |
| M. marinum M | MMAR_0490 | gabD1 | 0.0 | 80.74% (457) | succinate-semialdehyde dehydrogenase |
| M. avium 104 | MAV_4936 | gabD1 | 0.0 | 80.96% (457) | succinic semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6452 | - | 1e-104 | 43.81% (452) | [NADP+] succinate-semialdehyde dehydrogenase [Mycobacterium |
| M. thermoresistible (build 8) | TH_4137 | - | 1e-73 | 36.52% (460) | PUTATIVE putative [NADP+] succinate-semialdehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_1153 | gabD1 | 0.0 | 80.53% (457) | succinic semialdehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1844 | - | 1e-98 | 43.96% (455) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0490|M.marinum_M --------------------------------------------------
MUL_1153|M.ulcerans_Agy99 --------------------------------------------------
MAV_4936|M.avium_104 --------------------------------------------------
Mb0239c|M.bovis_AF2122/97 MRSVTCSATLVLPVIEPTPADRRPRHLLLGSAGHVSGRLDTGRFVQTHPA
Rv0234c|M.tuberculosis_H37Rv MRSVTCSATLVLPVIEPTPADRRPRHLLLGSAGHVSGRLDTGRFVQTHPA
MLBr_02573|M.leprae_Br4923 --------------------------------------------------
MAB_3471|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6452|M.smegmatis_MC2_155 -------------------------------------------------M
Mvan_1844|M.vanbaalenii_PYR-1 -------------------------------------------------M
Mflv_4860|M.gilvum_PYR-GCK -------------------------------------MTVASSWIDGAPV
TH_4137|M.thermoresistible__bu -------------------------------------MYIDGEWQSAASG
MMAR_0490|M.marinum_M ----MPIATTNPATGETVKTFTAASNDEVDAAIARAYARFQDYRRNTTLA
MUL_1153|M.ulcerans_Agy99 ----MPIATTNPATGETVKTFTAASNDEVDAAIARAYARFQDYRRNTTFA
MAV_4936|M.avium_104 ----MPIATINPATGETVKTFTPASDAEVDAAIARAYERFLDYRHSTTFA
Mb0239c|M.bovis_AF2122/97 KDVSVPIATINPATGETVKTFTAATDDEVDAAIARAHRRFADYRQ-TSFA
Rv0234c|M.tuberculosis_H37Rv KDVSVPIATINPATGETVKTFTAATDDEVDAAIARAHRRFADYRQ-TSFA
MLBr_02573|M.leprae_Br4923 ----MSIATINPATGETVKTFTPTTQDEVEDAIARAYARFDNYRH-TTFT
MAB_3471|M.abscessus_ATCC_1997 ----MPIATINPATGETIKTFTAASDAEIDAAIGRAHDRFRQYRT-TSFS
MSMEG_6452|M.smegmatis_MC2_155 NMTQ--YAVTDPATAEVVATYPTATDAEVQAAIEAAHKTGRTWAKSTTVA
Mvan_1844|M.vanbaalenii_PYR-1 TATQPPYRTLNPATGEVLATFDTATDDLIATAITAADDAYRAWRE-QPLE
Mflv_4860|M.gilvum_PYR-GCK KTGGAQHTVINPATGESVAEFALAQPGDVDRAVVSARAALPGWKG-ATPA
TH_4137|M.thermoresistible__bu KT----FASINPANGAVLDSVPDGEREDARRAIDAAAAALPDWRS-RTAY
:**.. : *: * : .
MMAR_0490|M.marinum_M QRAEWAHATADLIEAEADQTAALMTLEMGKTIASAKA-EVLKSAKGFRYY
MUL_1153|M.ulcerans_Agy99 QRAEWAHATADLIEAEADQTAALMTLEMGKTIASAKA-EVLKSAKGFRYY
MAV_4936|M.avium_104 QRAQWANATADLLEAEADEVAAMMTLEMGKTLKSAKA-EALKCAKGFRYY
Mb0239c|M.bovis_AF2122/97 QRARWANATADLLEAEADQAAAMMTLEMGKTLAAAKA-EALKCAKGFRYY
Rv0234c|M.tuberculosis_H37Rv QRARWANATADLLEAEADQAAAMMTLEMGKTLAAAKA-EALKCAKGFRYY
MLBr_02573|M.leprae_Br4923 QRAKWANATADLLEAEANQSAALMTLEMGKTLQSAKD-EAMKCAKGFRYY
MAB_3471|M.abscessus_ATCC_1997 DRAAWALSTADLLEAEADDVAALMTLEMGKTLASAKA-EVLKCAKGFRFY
MSMEG_6452|M.smegmatis_MC2_155 ERAALIRKVAELHEQRREQLADVIVREMGKPRDQSLG-EVDFCAAIYQYY
Mvan_1844|M.vanbaalenii_PYR-1 ARVDVVRHVGRLLRERATELAELAKLEMGKKLDEGTG-EVEFSASIFEFY
Mflv_4860|M.gilvum_PYR-GCK ERSAVLAKLAKLADDATADLVAEEVSQTGKPVRLATEFDVPGSVDNIDFF
TH_4137|M.thermoresistible__bu ERAEVLVEAHRIMLERRDDLAELMTREQGKPLRMART-EVGYAADFLLWF
* : : . : ** . :. .. ::
MMAR_0490|M.marinum_M ADNAAALLADEPADAGKVGASQAYTRYQPLGVVLAVMPWNFPLWQAVRFA
MUL_1153|M.ulcerans_Agy99 ADNAAALLADEPADAGKVGASQAYTRYQPLGVVLAVMPWNFPLWQAGRFA
MAV_4936|M.avium_104 AQNAEQLLADEPADAGKVGAARAYIRYQPLGVVLAVMPWNFPLWQAVRFA
Mb0239c|M.bovis_AF2122/97 AENAEALLADEPADAAKVGASAAYGRYQPLGVILAVMPWNFPLWQAVRFA
Rv0234c|M.tuberculosis_H37Rv AENAEALLADEPADAAKVGASAAYGRYQPLGVILAVMPWNFPLWQAVRFA
MLBr_02573|M.leprae_Br4923 AEKAETLLADEPAEAAAVGASKALARYQPLGVVLAVMPWNFPLWQTVRFA
MAB_3471|M.abscessus_ATCC_1997 AENAEAMLAPEPADASAVNASQAYAQYQPLGVVLAVMPWNFPLWQAVRFA
MSMEG_6452|M.smegmatis_MC2_155 ADNAEKFLADEPIEL-LDGEGSAVIKRSPIGVLLGIMPWNYPYYQVARFA
Mvan_1844|M.vanbaalenii_PYR-1 ADHAAALLADQRIPS-FSG-GTAVVQRRPVGVLLGVMPWNFPVYQVARFA
Mflv_4860|M.gilvum_PYR-GCK AGAARHLEG--KASAEYSGDHTSSIRREAVGVVATITPWNYPLQMAVWKV
TH_4137|M.thermoresistible__bu AEEAKRVYG--EVIPSARADQRFIVLAQPVGVCAAITPWNYPISMLTRKL
* * . . .:** : ***:*
MMAR_0490|M.marinum_M APALMAGNVGLLKHASNVPQSALYLADVIARGGFPDGCFQTLLVSASA--
MUL_1153|M.ulcerans_Agy99 APALMAGNVGLLKHASNVPQSALYLADVIARAGFPDGCFQTLLVSASA--
MAV_4936|M.avium_104 APALMAGNVGILKHASNVPQSALYLADVITRGGFPEGCFQTLLVPSSA--
Mb0239c|M.bovis_AF2122/97 APALMAGNVGLLKHASNVPQCALYLADVIARGGFPDGCFQTLLVSSGA--
Rv0234c|M.tuberculosis_H37Rv APALMAGNVGLLKHASNVPQCALYLADVIARGGFPDGCFQTLLVSSGA--
MLBr_02573|M.leprae_Br4923 APALMAGNVGLLKHASNVPQCALYLADVIARGGFPEDCFQTLLISANG--
MAB_3471|M.abscessus_ATCC_1997 APALMAGNVGLLKHASNVPQSALYLADVIARGGFPEGCFQTLLVPSSA--
MSMEG_6452|M.smegmatis_MC2_155 GPNLVVGNTILLKHAPQCPESAELLQLLFLEAGFPQGAYVDIRATNEQ--
Mvan_1844|M.vanbaalenii_PYR-1 APNLVLGNTILLKHAESVPQCALVIADVMRDAGVPEGVYTNLFASFDQ--
Mflv_4860|M.gilvum_PYR-GCK IPALAAGCSVVIKPAEITPLTTLTLARLASEAGLPDGVLNVVTGSGAD-V
TH_4137|M.thermoresistible__bu APALAAGCTSVLKPAEQTPLCAVETFKVFADAGVPAGVVNLVTCSDPEPV
* * * ::* * * : : .*.* . : .
MMAR_0490|M.marinum_M VEGILRDPRVAAATLTGSEPAGQSVGAIAGDEIKPTVLELGGSDPFIVMP
MUL_1153|M.ulcerans_Agy99 VEGILRDPRVAAATLTGSEPAGQSVGAIAGDEIKPTVLELGGSDPFIVMP
MAV_4936|M.avium_104 VERILRDPRVAAATLTGSEPAGQSVAAIAGDEIKPTVLELGGSDPFIVMP
Mb0239c|M.bovis_AF2122/97 VEAILRDPRVAAATLTGSEPAGQSVGAIAGNEIKPTVLELGGSDPFIVMP
Rv0234c|M.tuberculosis_H37Rv VEAILRDPRVAAATLTGSEPAGQSVGAIAGNEIKPTVLELGGSDPFIVMP
MLBr_02573|M.leprae_Br4923 VEAILRDPRVAAATLTGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMP
MAB_3471|M.abscessus_ATCC_1997 VERILRDPRVAAATLTGSEPAGRSVAAIAGDEVKPTVLELGGSDPFIVMP
MSMEG_6452|M.smegmatis_MC2_155 VANIIADPRVAAVSLTGSERAGAAVAEIAGRNLKKCVLELGGSDPFIVLS
Mvan_1844|M.vanbaalenii_PYR-1 VETIIGDPRVQGVSVTGSERAGAAVAATAGRSLKKCLLELGGSDPFIVLD
Mflv_4860|M.gilvum_PYR-GCK GAALAGHADVDMVTFTGSTPVGRRVMLAAAAHGHRTQLELGGKAPFVVFD
TH_4137|M.thermoresistible__bu ADELLGDPRVRKISFTGSTEVGRMIASRAGATMKRVSMELGGHAPFLVFP
: .. * :.*** .* : *. : :**** **:*:
MMAR_0490|M.marinum_M SADLDKAVSTAVTGRVQNNGQSCIAAKRFIAHADIYDAFVDKFVEQMSAL
MUL_1153|M.ulcerans_Agy99 SADLDKAVSTAVTGRVQNNGQSCIAAKRFIAHADIYDAFVDKFVEQMSAL
MAV_4936|M.avium_104 SADLDEAVKTAVTARVQNNGQSCIAAKRFIVHTDIYDTFVDKFVEQMKAL
Mb0239c|M.bovis_AF2122/97 SADLDAAVSTAVTGRVQNNGQSCIAAKRFIVHADIYDDFVDKFVARMAAL
Rv0234c|M.tuberculosis_H37Rv SADLDAAVSTAVTGRVQNNGQSCIAAKRFIVHADIYDDFVDKFVARMAAL
MLBr_02573|M.leprae_Br4923 SADLDAAVATAVTGRVQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETL
MAB_3471|M.abscessus_ATCC_1997 SADLEQAVSTAVTARVQNNGQSCIAAKRFIAHRDIYDDFLSRFVEKMAAL
MSMEG_6452|M.smegmatis_MC2_155 TDDLDATVQAAVDGRMENTGQACNAAKRFIVNESIYDDFLAKFTEKLLAA
Mvan_1844|M.vanbaalenii_PYR-1 -GDSKQLARTAWEFRVYNMGQACNSNKRMIVTDDLYDDFVAELVTLAGDL
Mflv_4860|M.gilvum_PYR-GCK DADLDAAIQGAVAGALINTGQDCTAATRAIVAEGLYDDFVAGVAEVMSKV
TH_4137|M.thermoresistible__bu DADPEHAAKGGAAVKFLNTGQACICPNRFIVHRSIAEPFIATLRARIEKL
* . . . * ** * . .* *. .: : *: .
MMAR_0490|M.marinum_M TVGDPTDPQTQVGPLATEQSRDEIAQQVDDAAAAGAVIRCGGKPLAGP--
MUL_1153|M.ulcerans_Agy99 TVGDPTDPQTQVGPLATEQSRDEIAQQVDDAAAAGAVIRCGGKPLAGP--
MAV_4936|M.avium_104 KVGDPTDPATDVGPLATESGRDEIAKQVDDAVAAGATLRCGGKPLDGP--
Mb0239c|M.bovis_AF2122/97 RVGDPTDPDTDVGPLATEQGRNEVAKQVEDAAAAGAVIRCGGKRLDRP--
Rv0234c|M.tuberculosis_H37Rv RVGDPTDPDTDVGPLATEQGRNEVAKQVEDAAAAGAVIRCGGKRLDRP--
MLBr_02573|M.leprae_Br4923 RVGDPTDPDTDVGPLATESGRAQVEQQVDAAAAAGAVIRCGGKRPDRP--
MAB_3471|M.abscessus_ATCC_1997 RVGDPTDPATDVGPLATEQGRDEVDKQVQQAVAAGASIRCGGTRPDGP--
MSMEG_6452|M.smegmatis_MC2_155 G--------EGIAPLSSEQAAVRLEDQVKSAVEAGAKLTAAGERKG----
Mvan_1844|M.vanbaalenii_PYR-1 AP-------AEIAPMSSRKAAETLAAQVADAVDKGATLHVGGELAEGP--
Mflv_4860|M.gilvum_PYR-GCK VIGDPADPDTDLGPLISAVHREKVAGMVERAPGEGGRVVTGGVAPDGP--
TH_4137|M.thermoresistible__bu VPGDGLAAGTTVGPLIDEVALKKMQRQVDDAVDKGAKLELGGSRLQGPAF
:.*: : * * *. : .*
MMAR_0490|M.marinum_M --GWYYPPTVITDITKDMNLYTEEVFGPVASVYRAADIDEAIEIANATTF
MUL_1153|M.ulcerans_Agy99 --GWYYPPTVITDITKDMNLYTEEVFGPVASVYRAADIDEAIEIANATTF
MAV_4936|M.avium_104 --GWFYPPTVVTDITKDMALYTEEVFGPVASMYRAADIDEAIEIANATTF
Mb0239c|M.bovis_AF2122/97 --GWFYPPTVITDISKDMALYTEEVFGPVASVFRAANIDEAVEIANATTF
Rv0234c|M.tuberculosis_H37Rv --GWFYPPTVITDISKDMALYTEEVFGPVASVFRAANIDEAVEIANATTF
MLBr_02573|M.leprae_Br4923 --GWFYPPTVITDITKDMALYTDEVFGPVASVYCATNIDDAIEIANATPF
MAB_3471|M.abscessus_ATCC_1997 --GWYYPPTVITDITKNMPIFGEEVFGPVASVYRATDIEEAIEVANATSF
MSMEG_6452|M.smegmatis_MC2_155 ---AFFPPAVLTGVTEDNPVYRQELFGPVAVVYKAGSEEEAVQIANDTPF
Mvan_1844|M.vanbaalenii_PYR-1 --AAFYTPAVLTGVTPDMRAYSEELFGPVAVVYRVRDEEEAVALANASAY
Mflv_4860|M.gilvum_PYR-GCK --GSFYRPTLIADVAESSEVWRDEIFGPVLTVRSFTDDDDAIRQANDTKY
TH_4137|M.thermoresistible__bu DNGHFYAPTILTGVTPDMLICSEETFGPIAPVMVFDDEDEAIAMANSTEY
:: *::::.:: . :* ***: : . ::*: ** : :
MMAR_0490|M.marinum_M GLGSNAWTQDEAEQRRFINDIEAGQVFINGMTVSYPELPFGGIKRSGYGR
MUL_1153|M.ulcerans_Agy99 GLGSNAWTQDEAEQRRFINDIEAGQVFINGMTVSYPELPFGGIKRSGYGR
MAV_4936|M.avium_104 GLGSNAWTNDAAEQQRFIDDIEAGQVFINGMTVSYPELGFGGVKRSGYGR
Mb0239c|M.bovis_AF2122/97 GLGSNAWTRDETEQRRFIDDIVAGQVFINGMTVSYPELPFGGVKRSGYGR
Rv0234c|M.tuberculosis_H37Rv GLGSNAWTRDETEQRRFIDDIVAGQVFINGMTVSYPELPFGGVKRSGYGR
MLBr_02573|M.leprae_Br4923 GLGSNAWTQNESEQRHFIDNIVAGQVFINGMTVSYPELPFGGIKRSGYGR
MAB_3471|M.abscessus_ATCC_1997 GLGANAWTRETAEQQRFINELDAGQVFINGMTVSYPEVPFGGIKRSGYGR
MSMEG_6452|M.smegmatis_MC2_155 GLGSYVFTTDSEQAARVADKIEAGMVFVNAVGAEGAELPFGGVKRSGFGR
Mvan_1844|M.vanbaalenii_PYR-1 GLGASVFSTDTARAEKVAQRLEAGMVNVNTPAGEGPDIPFGGVKRSGFGR
Mflv_4860|M.gilvum_PYR-GCK GLAASAWTRDVYRAQRASREIHAGCVWINDHIPIISEMPHGGFGASGFGK
TH_4137|M.thermoresistible__bu GLAGYIYTRDISRAIRVGEALDFGMIGINDINPTSAAVPFGGIKQSGLGR
**.. :: : . : : * : :* . : .**. ** *:
MMAR_0490|M.marinum_M ELAGHGIREFCNIKTVWVG-----------------
MUL_1153|M.ulcerans_Agy99 ELAGHGIREFCNIKTVWVG-----------------
MAV_4936|M.avium_104 ELAGLGIRAFCNAKTVWIGSSKSGDAGGGSKVE---
Mb0239c|M.bovis_AF2122/97 ELSAHGIREFCNIKTVWIA-----------------
Rv0234c|M.tuberculosis_H37Rv ELSAHGIREFCNIKTVWIA-----------------
MLBr_02573|M.leprae_Br4923 ELSTHGIREFCNIKTVWIA-----------------
MAB_3471|M.abscessus_ATCC_1997 ELSAHGIREFCNIKTVWIA-----------------
MSMEG_6452|M.smegmatis_MC2_155 ELGRYGIDEFVNKKLIRVVD----------------
Mvan_1844|M.vanbaalenii_PYR-1 ELGALGIDEFVNKQVYVVAD----------------
Mflv_4860|M.gilvum_PYR-GCK DMSQYSLEEYLSIKHVMSDITSAVDKDWHRTIFTKR
TH_4137|M.thermoresistible__bu EGARAGIEEYLDTKVLGIAL----------------
: . .: : . :