For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSAGPREGETEPPPVPRPDFQSPEGRRQLHRAIAASAMGNATEWYDYGVYAATTTFLTQAFFPTLGTAFT MLGYAISFLLRPLGGMVWGPLGDRLGRKSVMAITIVLMALSTTLIGILPSWATIGMAAPALLILLRVIQG FSTGGEYGGAATFMAEFAPDKKRGKYGSFLEFGTLGGFAVGAAFVLVLDATLSDDAMHTWGWRIPFLVAL PMGLIGWYLRSRLDDPPLFKEMQEQEPEGRETAFERLRDLVRDYRRPMVVMMLLVIALNITNYTLLSYQP TYLKTRLGMSETQGQIVILIGEVAMMAFLPFCGALSDRVGRKPMWAFSLTGLLVLSLPMFWLMGQGFGWA IVGFAVLGLLYIPQLSTITATFPCMFPTQVRYAGFAISYNLSTAAFGGTAPLVNDLVVARTGWDLFPAAY LMVACVVGLCALPFLIETAGASIAGTEIPCNEAAVPTDTVRA*
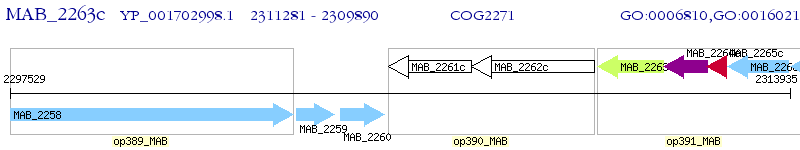
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2263c | - | - | 100% (463) | major facilitator transporter |
| M. abscessus ATCC 19977 | MAB_4485 | - | 4e-66 | 34.11% (428) | putative transporter |
| M. abscessus ATCC 19977 | MAB_3808c | - | 5e-57 | 32.33% (433) | dicarboxylic acid transport integral membrane protein KgtP |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3503c | kgtP | 6e-56 | 32.18% (435) | dicarboxylic acid transport integral membrane protein KgtP |
| M. gilvum PYR-GCK | Mflv_0953 | - | 1e-145 | 57.30% (445) | general substrate transporter |
| M. tuberculosis H37Rv | Rv3476c | kgtP | 5e-56 | 32.41% (435) | dicarboxylic acid transport integral membrane protein KgtP |
| M. leprae Br4923 | MLBr_01562 | - | 3e-07 | 26.09% (161) | putative transmembrane efflux protein |
| M. marinum M | MMAR_4238 | - | 3e-53 | 31.93% (429) | integral membrane transport protein |
| M. avium 104 | MAV_4627 | - | 2e-59 | 32.05% (440) | major facilitator superfamily protein |
| M. smegmatis MC2 155 | MSMEG_1477 | - | 1e-146 | 55.63% (462) | major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_3400 | - | 1e-151 | 58.67% (450) | PUTATIVE General substrate transporter |
| M. ulcerans Agy99 | MUL_0949 | - | 1e-53 | 31.93% (429) | integral membrane transport protein |
| M. vanbaalenii PYR-1 | Mvan_5839 | - | 1e-145 | 59.44% (429) | general substrate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0953|M.gilvum_PYR-GCK ---MDSR------------DARPPTIEKEQ-----PPAVLKKAIAASA--
Mvan_5839|M.vanbaalenii_PYR-1 --------------------MGMPTTHEDP-----PPGVLKKAISASA--
MSMEG_1477|M.smegmatis_MC2_155 ---MDSKEADASKPSQADADQQAPTAHAEEVKA--PPEVVKRAVAASA--
TH_3400|M.thermoresistible__bu ---LTTERTD---------PPQDPSVEQEQ-----PPGVLRKAIAASA--
MAB_2263c|M.abscessus_ATCC_199 ---MSSAGPR-------EGETEPPPVPRPDFQSPEGRRQLHRAIAASA--
Mb3503c|M.bovis_AF2122/97 -----------------MTVSIAPPSRPSQ----AETRRAIWNTIRGS--
Rv3476c|M.tuberculosis_H37Rv -----------------MTVSIAPPSRPSQ----AETRRAIWNTIRGS--
MMAR_4238|M.marinum_M ---------------------------------------MSRVALACV--
MUL_0949|M.ulcerans_Agy99 ---------------------------------------MSRVALACV--
MAV_4627|M.avium_104 ----------MAATLPAPAPAGLADGHPLDESAEQQRRRLRIVVAASL--
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYPTLLPSGRF
Mflv_0953|M.gilvum_PYR-GCK -----IGNATEWFDYGIYAYGVTYISAAIFPG--DAANATLLALMTFAVS
Mvan_5839|M.vanbaalenii_PYR-1 -----IGNATEWFDYGIYAYGVSYISAAIFPG--DAANATLLALMTFAVS
MSMEG_1477|M.smegmatis_MC2_155 -----VGNFTEWFDYGIYAYGVSYISAAIFPG--DGKSATLLALMTFAVS
TH_3400|M.thermoresistible__bu -----VGNATEWFDYGLYAYGVTYISVAIFPG--DGATATLLALMTFAVS
MAB_2263c|M.abscessus_ATCC_199 -----MGNATEWYDYGVYAATTTFLTQAFFP-----TLGTAFTMLGYAIS
Mb3503c|M.bovis_AF2122/97 -----SGNLVEWYDVYVYTVFATYFEDQFFDR--ADRNSTVYVYAIFAVT
Rv3476c|M.tuberculosis_H37Rv -----SGNLVEWYDVYVYTVFATYFEDQFFDR--ADRNSTVYVYAIFAVT
MMAR_4238|M.marinum_M -----VGSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MUL_0949|M.ulcerans_Agy99 -----VGSTVEYYDFCIYGTAAALVFPAVFYPGLSPALATIASMGTFATA
MAV_4627|M.avium_104 -----LGTTVEWYDFFLYATAAGLVFNKVFFPNESSLVGTLLAFATFAVG
MLBr_01562|M.leprae_Br4923 PSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGITA
. . : . : :
Mflv_0953|M.gilvum_PYR-GCK FLVRPLGGFVWG-PLGDRLGRKRVLAMTILLMAGATFCVGLVPTYAAIGM
Mvan_5839|M.vanbaalenii_PYR-1 FLVRPLGGFVWG-PLGDRLGRKRVLAITIVLMAGATFCVALVPTYDMIGM
MSMEG_1477|M.smegmatis_MC2_155 FLVRPLGGLVWG-PLGDRIGRKQVLAITILLMAGATLCVGLVPPYATIGV
TH_3400|M.thermoresistible__bu FLVRPLGGFVWG-PLGDRLGRRQVLAITILLMAGATFCVGLVPSYDSIGL
MAB_2263c|M.abscessus_ATCC_199 FLLRPLGGMVWG-PLGDRLGRKSVMAITIVLMALSTTLIGILPSWATIGM
Mb3503c|M.bovis_AF2122/97 FVTRPVGSWFFG-RFADRRGRRAALTFSVSLMAACSLIVALVPSRSSIGV
Rv3476c|M.tuberculosis_H37Rv FVTRPVGSWFLG-RFADRRGRRAALTFSVSLMAACSLIVALVPSRSSIGV
MMAR_4238|M.marinum_M FLSRPLGAAVFG-HFGDRLGRKKTLVATLLIMALSTVTVGVVPSTATIGV
MUL_0949|M.ulcerans_Agy99 FLSRPLGAAVFG-HFGDRLGRKKTLVATLLIMALSTVTVGVVPSTATIGV
MAV_4627|M.avium_104 FVMRPIGGLVFG-HIGDRIGRKRSLALTMLIMGGATALIGVLPTAAQIGA
MLBr_01562|M.leprae_Br4923 YVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVA--------
:: * . * :.* **: : : :: .: .:
Mflv_0953|M.gilvum_PYR-GCK WAPFLLVLLRMIQGFSTGGEYGG-----AATFMAEYSPSRRRGLLGSFLE
Mvan_5839|M.vanbaalenii_PYR-1 WAPFLLVLLRMIQGFSTGGEYGG-----AATFMAEYAPNRRRGLLGSFLE
MSMEG_1477|M.smegmatis_MC2_155 AAPILMVVLRMIQGFSTGGEYGG-----AATFMAEYSPSRRRGLFGSFLE
TH_3400|M.thermoresistible__bu WAPVLMVILRMIQGFSTGGEYGG-----AATFMAEYAPSRRRGLLGSFLE
MAB_2263c|M.abscessus_ATCC_199 AAPALLILLRVIQGFSTGGEYGG-----AATFMAEFAPDKKRGKYGSFLE
Mb3503c|M.bovis_AF2122/97 AAPILLILCRLVQGFATGGEYGT-----SVTYMSEAATRERRGYFSSFQY
Rv3476c|M.tuberculosis_H37Rv AAPILLILCRLVQGFATGGEYGT-----SATYMSEAATRERRGYFSSFQY
MMAR_4238|M.marinum_M SAPLILVALRLLQGFAVGGEWAG-----SALLSIESAPAHKRGYYGMFTV
MUL_0949|M.ulcerans_Agy99 SAPLILVALRLLQGFAVGGEWAG-----SALLSIESAPAHKRGYYGMFTV
MAV_4627|M.avium_104 WAPVLLLVLRVLQGFALGGEWGG-----AVLLAVEHSPGDRRGRYGAVPQ
MLBr_01562|M.leprae_Br4923 WDEVTLVIARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTA
:: *::**:. . :. : . . .
Mflv_0953|M.gilvum_PYR-GCK FGTLAGFSCGALLMLG-------CSLVLG--DEDMQAWGWRLPFLVAAPL
Mvan_5839|M.vanbaalenii_PYR-1 FGTLGGFSLGALLMLG-------CSLVLG--DEQMHAWGWRLPFLVAAPL
MSMEG_1477|M.smegmatis_MC2_155 FGTLAGFSFGALLMLG-------FSLLLS--DDQMGSWGWRLPFLVAAPL
TH_3400|M.thermoresistible__bu FGTLAGFSLGALMMLG-------VSLMLS--DDQMNTWGWRVPFLIAAPL
MAB_2263c|M.abscessus_ATCC_199 FGTLGGFAVGAAFVLV-------LDATLS--DDAMHTWGWRIPFLVALPM
Mb3503c|M.bovis_AF2122/97 VTLVGGHVLAQFTLLV-------ILAVFT--REQVHEFGWRIGFAVGGGA
Rv3476c|M.tuberculosis_H37Rv VTLVGGHVLAQFTLLV-------ILAVFT--REQVHEFGWRIGFAVGGGA
MMAR_4238|M.marinum_M LGGGVALVVSGLTFLG-------VNYTIGESSSAFLQWGWRVPFLLSALL
MUL_0949|M.ulcerans_Agy99 LGGGVALVVSGLTFLG-------VNYTIGESSSALLQWGWRVPFLLSALL
MAV_4627|M.avium_104 VGLALGLALGTGIFAF-------LQIVLG--PARFLSYGWRIGFLLSVLL
MLBr_01562|M.leprae_Br4923 IGSVMGLVVGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKERMKL
. . . : . . : .
Mflv_0953|M.gilvum_PYR-GCK GLIGVYLRSRLEDTPVYRE--LEQEGRT-EESTSMQFRDLVV--------
Mvan_5839|M.vanbaalenii_PYR-1 GLIGLYLRSRLEDTPVFRE--LEAKGGT-EPETTTQFRDLLG--------
MSMEG_1477|M.smegmatis_MC2_155 GLIGVYLRSRLEDTPIYRE--LEESGES-EEHPTGEFKALVV--------
TH_3400|M.thermoresistible__bu GLIGLYLRSRLDDTPIFRE--LQESDRE-EKSIWTEFKDLLV--------
MAB_2263c|M.abscessus_ATCC_199 GLIGWYLRSRLDDPPLFKE--MQEQEPEGRETAFERLRDLVR--------
Mb3503c|M.bovis_AF2122/97 AIVVFWLRRTMDESLSQER--LTAIKAG-RDHDSGSLRELAT--------
Rv3476c|M.tuberculosis_H37Rv AIVVFWLRRTMDESLSQER--LTAIKAG-RDHDSGSLRELAT--------
MMAR_4238|M.marinum_M IAFALYVRLEINETPVFARAMAQADDQESTAAARAPLAAVLC--------
MUL_0949|M.ulcerans_Agy99 IAFALYVRLEINETPVFARAMAQADDQESTAAARAPLAAVLC--------
MAV_4627|M.avium_104 VAIGIVVRLRVEETPAFR------ELQDLQAASTVPIRDILRER------
MLBr_01562|M.leprae_Br4923 DAAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVE
: . : :
Mflv_0953|M.gilvum_PYR-GCK ------------RYWRPILQMGGLVVALNVVNYTLLTYMPTYLENSIGLS
Mvan_5839|M.vanbaalenii_PYR-1 ------------RYWRPILQLGGLVVALNVVNYTLLSYMPTYLENRIGLS
MSMEG_1477|M.smegmatis_MC2_155 ------------DYWSPILRLGGLVVALNVVNYTLLTYMPTYLESAIGLS
TH_3400|M.thermoresistible__bu ------------EYRGQILRLGGLVVALNVVNYTLLSYMPTYLQTSIGLS
MAB_2263c|M.abscessus_ATCC_199 ------------DYRRPMVVMMLLVIALNITNYTLLSYQPTYLKTRLGMS
Mb3503c|M.bovis_AF2122/97 ------------HYWKPLLLCFLVTLGGTVAFYTYSVNAPAIVKSVYGSQ
Rv3476c|M.tuberculosis_H37Rv ------------HYWKPLLLCFLVTLGGTVAFYTYSVNAPAIVKSVYGSQ
MMAR_4238|M.marinum_M ------------RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYS
MUL_0949|M.ulcerans_Agy99 ------------RQRREIVLAAGAMLAAFGCLYLASTFLPSYAQLHLGYS
MAV_4627|M.avium_104 ------------RSRRNTVLGLLSRWAEGSAFNTWGVFAISYATGALGLN
MLBr_01562|M.leprae_Br4923 RTAENPVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYS
: * .
Mflv_0953|M.gilvum_PYR-GCK ADQSLMVPIIGMLAMMVFVPFAGLLSDRVGRKP-LWWFSLIGLFVAGIPA
Mvan_5839|M.vanbaalenii_PYR-1 PDQSLIVPVIGMLSMMVFVPFAGLLSDRVGRKP-LWWFSLIGLFVAGVPM
MSMEG_1477|M.smegmatis_MC2_155 TDQSLVVPIIGMLSMMVFLPFAGRISDRVGRKP-MWWFSVVGLFVAGVPM
TH_3400|M.thermoresistible__bu TDTALVVPIIGMLSMMVFLPFAGLLSDRFGRKP-MWWISLGGLLALVVPM
MAB_2263c|M.abscessus_ATCC_199 ETQGQIVILIGEVAMMAFLPFCGALSDRVGRKP-MWAFSLTGLLVLSLPM
Mb3503c|M.bovis_AF2122/97 AMTATWINLVGLILLMMLQPIGGMISDKIGRKPLLLWFGVGGLIYTYVLV
Rv3476c|M.tuberculosis_H37Rv AMTATWINLVGLILLMMLQPIGGMISDKIGRKPLLLWFGVGGLIYTYVLV
MMAR_4238|M.marinum_M RNLVLLIGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVM
MUL_0949|M.ulcerans_Agy99 RNLVLLIGVLGGLVCIGFVALSAILCDRFGRRRVMLVGWAVGLFWSPLVM
MAV_4627|M.avium_104 RVSVLIAVTIAALLMAVLLPVSGVLTDRFGARRVYLAGIACYGLAAFPAF
MLBr_01562|M.leprae_Br4923 ALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGW
: . . : .:.. : .
Mflv_0953|M.gilvum_PYR-GCK FLLMG------TNLAGAVIGFAILGLLYVPQLATISA-------------
Mvan_5839|M.vanbaalenii_PYR-1 FLLMG------TNLWGAVIGFAVLGLLYVPQLATISA-------------
MSMEG_1477|M.smegmatis_MC2_155 FMLMS------TNLVGAIIGFAVLGLLYVPQLATISA-------------
TH_3400|M.thermoresistible__bu FMLMS------TSVPGAIIGFAVLGLLYVPQLATISA-------------
MAB_2263c|M.abscessus_ATCC_199 FWLMG------QGFGWAIVGFAVLGLLYIPQLSTITA-------------
Mb3503c|M.bovis_AF2122/97 TYLPE------TRSPTMSFLLVAVGYVILTGYCSINA-------------
Rv3476c|M.tuberculosis_H37Rv TYLPE------TRSPTMSFLLVAVGYVILTGYCSINA-------------
MMAR_4238|M.marinum_M PLIGS------GDTVGFTVALLGLY-AVAASGFGPAA-------------
MUL_0949|M.ulcerans_Agy99 PLIGS------GDTVGFTVALLGLY-AVAASGFGPAA-------------
MAV_4627|M.avium_104 ALFGT------KKLLWFGLAMVIVFGVVHALFYGAQG-------------
MLBr_01562|M.leprae_Br4923 AFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAI
: . : . .
Mflv_0953|M.gilvum_PYR-GCK TFP-AMFPTQVRYAGFAIAYNVSTSIFGGTAP----AVNDWLVGATGDNL
Mvan_5839|M.vanbaalenii_PYR-1 TFP-AMFPTQVRYAGFAIAYNVSTSLFGGTAP----AINQWLTGETGDLL
MSMEG_1477|M.smegmatis_MC2_155 TFP-AMFPTQVRFAGFAIAYNVSTSIFGGTAP----AINDWLVNLTGDNL
TH_3400|M.thermoresistible__bu TFP-AMFPTHVRYAGFAIAYNVSTSLFGGTAP----AANDWLIQQTGNNL
MAB_2263c|M.abscessus_ATCC_199 TFP-CMFPTQVRYAGFAISYNLSTAAFGGTAP----LVNDLVVARTGWDL
Mb3503c|M.bovis_AF2122/97 LVKSELFPAHVRALGVGVGYALANSVFGGTAP----LIYQALKERDQVPM
Rv3476c|M.tuberculosis_H37Rv LVKSELFPAHVRALGVGVGYALANSVFGGTAP----LIYQALKERDQVPM
MMAR_4238|M.marinum_M SLIPELFATRYRYTGTALALNVAGIVGGAAPP----LIAGTLLASYGSSA
MUL_0949|M.ulcerans_Agy99 SLIPELFATRYRYTGTALALNVAGIVGGAAPP----LIAGTLLASYGSSA
MAV_4627|M.avium_104 TLYSALYPTNTRYTGLSFVYQFSGVYASGVTP----MILTALIAALGG-A
MLBr_01562|M.leprae_Br4923 ALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGYT
. .... :
Mflv_0953|M.gilvum_PYR-GCK VPAYYMMGACVIGALALIRIPETTRCPLNGTATPGTDEAPAPVEYEKSSA
Mvan_5839|M.vanbaalenii_PYR-1 FPAYYMMGACVIGAIALIKVPETARCPIGGTVTPGTEEAADPVPFEKQNA
MSMEG_1477|M.smegmatis_MC2_155 VPAYYMMVACVIGAISLVRMPETARCPINGTEIPGTPEAPPQLDYKEPAH
TH_3400|M.thermoresistible__bu MPAFYMMAACLVGMLALVKLPETARCPIGGTEIPGTEDAPPPVEYDLVGN
MAB_2263c|M.abscessus_ATCC_199 FPAAYLMVACVVGLCALPFLIETAGASIAGTEIP-CNEAAVPTDTVRA--
Mb3503c|M.bovis_AF2122/97 FIAYVTACIAVSLIVYVFFIKNKADTYLDREQGFAFYGHA----------
Rv3476c|M.tuberculosis_H37Rv FIAYVTACIAVSLIVYVFFIKNKADTYLDREQGFAFYGHA----------
MMAR_4238|M.marinum_M VGLMMTALVAAS-LLCTYLLPETAGAPLTAA-------------------
MUL_0949|M.ulcerans_Agy99 VGLMMTALVAAS-LLCTYLLPETAGAPLTAA-------------------
MAV_4627|M.avium_104 PWLACGYLVATA-VISVLATALIRDRDLYL--------------------
MLBr_01562|M.leprae_Br4923 YGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL--------
Mflv_0953|M.gilvum_PYR-GCK --
Mvan_5839|M.vanbaalenii_PYR-1 --
MSMEG_1477|M.smegmatis_MC2_155 AS
TH_3400|M.thermoresistible__bu R-
MAB_2263c|M.abscessus_ATCC_199 --
Mb3503c|M.bovis_AF2122/97 --
Rv3476c|M.tuberculosis_H37Rv --
MMAR_4238|M.marinum_M --
MUL_0949|M.ulcerans_Agy99 --
MAV_4627|M.avium_104 --
MLBr_01562|M.leprae_Br4923 --