For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RSIDTDSVAPAVPKPPVPRLPPRLPIHKAALLGLSCLGDPTRLPYLLLPKIANRYGDIVRLFTGPTPALT LTLINHPDYVDHVFTRHHDRYVKHEATIELVSGEPVALPLLEGREWKRVRSAFNPYFGERALAQATPLMM EGITERVDAWSRHVHSGELVDLEHELGAVVMDGLMRSMFKVRLRPAEIDHAVDGARRYGVYVISRVAMHF LPRWLPNPLRRSGEEAKAELFGILDRFVQERAGCPSTGTPDLVDTLLALEFDGCPQIRERRRRSEAAGLV FAGFETTAAALAWTIALLCRNPIALGKAYAEVDALGGKTLAYDDLENLKYLRACFDEAQRFQAAPANVRT AIEDDEVGGYFIPRGSQVIITQYALQRDPRFWNEPERFNPDRFLTDRINRNTFLPFSIGPRKCMGTRMAY IEGTLVLGAILQRYAFQIRDGWTPRHRVRVSTGLAGGLPARLFAR*
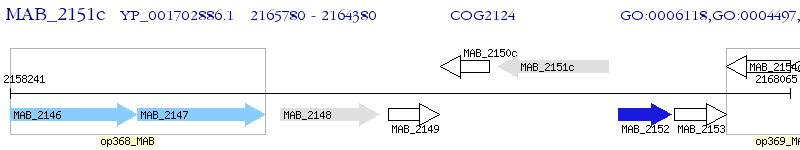
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2151c | - | - | 100% (466) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_4944c | - | 2e-72 | 35.45% (426) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_3275 | - | 1e-63 | 34.35% (457) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1429c | cyp132 | 5e-37 | 30.64% (408) | cytochrome P450 132 |
| M. gilvum PYR-GCK | Mflv_0056 | - | 4e-32 | 29.58% (382) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv1394c | cyp132 | 9e-37 | 30.88% (408) | cytochrome P450 132 |
| M. leprae Br4923 | MLBr_02088 | - | 3e-12 | 26.96% (204) | putative cytochrome p450 |
| M. marinum M | MMAR_0281 | cyp183B1 | 2e-27 | 27.42% (445) | cytochrome P450 183B1 Cyp183B1 |
| M. avium 104 | MAV_3106 | - | 5e-25 | 28.49% (372) | P450 heme-thiolate protein |
| M. smegmatis MC2 155 | MSMEG_0822 | - | 3e-29 | 27.66% (394) | cytochrome P450 |
| M. thermoresistible (build 8) | TH_3255 | - | 6e-27 | 27.35% (373) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_0604 | cyp185A4 | 1e-23 | 26.05% (334) | cytochrome P450 185A4 Cyp185A4 |
| M. vanbaalenii PYR-1 | Mvan_0245 | - | 1e-30 | 29.85% (335) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0056|M.gilvum_PYR-GCK ---------MVAGTEPG-VGHLPLAPKNPLPYRRLLTLVRALDTGQL---
TH_3255|M.thermoresistible__bu ---------MVVTTERADVATLPPAPRNPLPYWRQVRAVRSFHTGQE---
Mvan_0245|M.vanbaalenii_PYR-1 ---------MVGTVEREDLGALPLAPRNPLPYRQQVRAIRTFHTGLE---
MUL_0604|M.ulcerans_Agy99 ------------MVDPPDIRSLPLAPRNPLPYWQQLKALKTLHVGSE---
MSMEG_0822|M.smegmatis_MC2_155 ---------MVETASPP-LTELPLAPRNTLPCREQIRCLRSFIDGMQ---
Mb1429c|M.bovis_AF2122/97 ----------MATATTQRPLKGPAKRMSTWTMTREAITIGFDAGDGF---
Rv1394c|M.tuberculosis_H37Rv ----------MATATTQRPLKGPAKRMSTWTMTREAITIGFDAGDGF---
MMAR_0281|M.marinum_M ----------MPTSIPIAPAALPLLGHTLPLLRNPLSFLLSLPARGD---
MAV_3106|M.avium_104 -----------------------------MRYRPGEALLALYRRRGP---
MAB_2151c|M.abscessus_ATCC_199 -------MRSIDTDSVAPAVPKPPVPRLPPRLPIHKAALLGLSCLGDPTR
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTR---
:
Mflv_0056|M.gilvum_PYR-GCK -------QVRAAGGPITRVQVAPRWLFPPFVAVFSPRGVRDVLGRNDAFS
TH_3255|M.thermoresistible__bu -------LLRDAGGPVK-------WAIPPIVLVTSPAGIRDVLNRRDALS
Mvan_0245|M.vanbaalenii_PYR-1 -------TLRDAGGPVTRLKLAPKWLMPPMVVATSPQGARDILARGGGHI
MUL_0604|M.ulcerans_Agy99 -------ALRDAGGSVTRIWLGPKRLVPPMVMVTSPQGARDVLGRSGALS
MSMEG_0822|M.smegmatis_MC2_155 -------RLRDVGGPVTRIVLGPRWLVPPALLVTSPQGAHDVLSLPDAVA
Mb1429c|M.bovis_AF2122/97 -------LGRLRGSDITRFRCAGRRFVS----ISHPDYVDHVLHEARLKY
Rv1394c|M.tuberculosis_H37Rv -------LGRLRGSDITRFRCAGRRFVS----ISHPDYVDHVLHEARLKY
MMAR_0281|M.marinum_M -------LVRIRIGPAT------------AVVACTAELTHEILCDDRIFD
MAV_3106|M.avium_104 -------VIDAGAGRRG------------YTLLLGAEANKFVFANADAFS
MAB_2151c|M.abscessus_ATCC_199 LPYLLLPKIANRYGDIVRLFTGPTPALT-LTLINHPDYVDHVFTRHHDRY
MLBr_02088|M.leprae_Br4923 -------ADPFPVYRALIDYGPMQLPGMPLTVFSSFSDCDEALRHPLSAS
:
Mflv_0056|M.gilvum_PYR-GCK ER--CIVHEEVRDMAG-------DSLFVLPNEPWRPRKRALQPVFTPQNV
TH_3255|M.thermoresistible__bu ER--CQVHHEVRHLGG-------DSLFVLPTDPWRARRRALAPVFTPTAV
Mvan_0245|M.vanbaalenii_PYR-1 DK--TRVHHEMRRLLG-------ANLFDLTHEPWLPRRRALQPIFTKQHV
MUL_0604|M.ulcerans_Agy99 DRGTLPLLKDLRRAIG-------ESVLNLPHDTWLPRRRTLQPIFTKQRV
MSMEG_0822|M.smegmatis_MC2_155 DRGGSRNMRQLQRLMG-------GNLLDLPHDRWLPRRRTLQPLFTKKHV
Mb1429c|M.bovis_AF2122/97 VK--SDEYGPIRATAG-------LNLLTDEGDSWARHRGALNSTFARRHL
Rv1394c|M.tuberculosis_H37Rv VK--SDEYGPIRATAG-------LNLLTDEGDSWARHRGALNSTFARRHL
MMAR_0281|M.marinum_M KG--GIFFDRFREMLG-------DGLGTCPRSQHRRQRRLLQPAFHTQRL
MAV_3106|M.avium_104 WR---ATFENLALVDGP------TALIVSDGDDHRRRRSVVAPGLRHRQI
MAB_2151c|M.abscessus_ATCC_199 VK----HEATIELVSGEP-----VALPLLEGREWKRVRSAFNPYFGERAL
MLBr_02088|M.leprae_Br4923 DRLKATLAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVV
. . : . : :
Mflv_0056|M.gilvum_PYR-GCK RRFGGHMSRAAEVFVDAWR----DGGDVDLDLECRRITMQSLGRSVLGVD
TH_3255|M.thermoresistible__bu RSYGGYMSEAAQRVADGWQ----AAREVDLDAECRRLTMRSLGRSVLGLD
Mvan_0245|M.vanbaalenii_PYR-1 REFAGHMATAAQTVADSWA----DGTELDLDTECRKLTLRALGRSVLGLD
MUL_0604|M.ulcerans_Agy99 AGFAGHMSEAANHILLGWP----DGAEVDLNAASHALVLRVVGHSVFGLD
MSMEG_0822|M.smegmatis_MC2_155 PRFAGHMAAAAQSVAVSWA----DGATVDLDRACRALTLRALGRSVFGVD
Mb1429c|M.bovis_AF2122/97 RGLVGLMIDPIADVTAALV----PGAQFDMHQSMVETTLRVVANALFSQD
Rv1394c|M.tuberculosis_H37Rv RGLVGLMIDPIADVTAARV----PGAQFDMHQSMVETTLRVVANALFSQD
MMAR_0281|M.marinum_M PGYAQTMTTEVAATVGRWQ----NGQIIDVPAEMMAITGRTLLATMFSNS
MAV_3106|M.avium_104 QDYVTTMVSCIDRVIDGWR----PGQRLDVYQHCRAAVRRSTAESLFGRR
MAB_2151c|M.abscessus_ATCC_199 AQATPLMMEGITERVDAWSRHVHSGELVDLEHELGAVVMDGLMRSMFKVR
MLBr_02088|M.leprae_Br4923 QALEGDIAALVDSLLDKGA----AAGQFDVIADLAFPLAVAVICRLLGVP
: . .*: ::
Mflv_0056|M.gilvum_PYR-GCK LNESADVIAEHMH----------------VASSYTADRALRPVRAPRWLP
TH_3255|M.thermoresistible__bu INERADAISEPMH----------------IASSYTADRALRPVRAPRWLP
Mvan_0245|M.vanbaalenii_PYR-1 LDEHSDAIAEPLR----------------TALEYIADRALQPISAPRWLP
MUL_0604|M.ulcerans_Agy99 LQDKADVITTGVT----------------LAAKWAADRSARPVRAPHWLP
MSMEG_0822|M.smegmatis_MC2_155 LDQRAEDVGPALR----------------TSLSWVADRASRPVNLPQWVP
Mb1429c|M.bovis_AF2122/97 FGPLVQSMHDLATRGLRRAEKLERLGLWGLMPRTVYDTLIWCIYSGVHLP
Rv1394c|M.tuberculosis_H37Rv FGPLVQSMHDLATRGLRRAEKLERLGLWGLMPRTVYDTLIWCIYSGVHLP
MMAR_0281|M.marinum_M LPNPALRQALDDL----------------TAIFAVLYWRMLTPPPLDRLP
MAV_3106|M.avium_104 LAVHSDALGEYLQ-----------------PLLDLTHQPPQLVGLQRRIN
MAB_2151c|M.abscessus_ATCC_199 LRPAEIDHAVDGAR----------------RYGVYVISRVAMHFLPRWLP
MLBr_02088|M.leprae_Br4923 YEDAPEFG----------------------RVSALLVQSVDPFITITGEP
Mflv_0056|M.gilvum_PYR-GCK TPARRRAAHAVAEMRQVAADMLQVCRDDPDRDAPLVRALMDARDPET-GM
TH_3255|M.thermoresistible__bu TPARRRARAAVATMRRITDEMLQACRADPDRDAPLVHALIRATDPET-GR
Mvan_0245|M.vanbaalenii_PYR-1 TPARRRARAASSTLHKLADKILQACRTDPTRDAPLVQALIAASDPAT-GR
MUL_0604|M.ulcerans_Agy99 TPARPRARGAYDAMYKIAADMLAACRADPSREAPLITALIDATGPNT-GE
MSMEG_0822|M.smegmatis_MC2_155 TRGQRKARAGNATLHRLAAEILADVRADPDRDAPLVRALLEARDPET-GR
Mb1429c|M.bovis_AF2122/97 PPLR-EMQEITLTLDRAINSVIDRRLAEPTNSADLLNVLLSAD-----GG
Rv1394c|M.tuberculosis_H37Rv PPLR-EMQEITLTLDRAINSVIDRRLAEPTNSADLLNVLLSAD-----GG
MMAR_0281|M.marinum_M TVGNRRYHRASARLRHTIGSAVAGRRAAGVDRGDLLSAILASHEVDSSTV
MAV_3106|M.avium_104 APAWRRAMAARQRINNLVDTLIAEARAAPNPDDHMLTMLIDGRGDEG--Y
MAB_2151c|M.abscessus_ATCC_199 NPLRRSGEEAKAELFGILDRFVQERAGCPSTGTPDLVDTLLALEFDG-CP
MLBr_02088|M.leprae_Br4923 PEATEERLRAGVWLRDYLEQLVKCRRGTPGED--LISRLIELDESGD---
: : : :
Mflv_0056|M.gilvum_PYR-GCK ALSDDDICNDLLIFMLAGHDTTATALTYALWVLGHHPDVQDRVAAEAAAL
TH_3255|M.thermoresistible__bu GLSDEDICNDLLIFMLAGHDTTATALTYALWALGHHPEIQDRVAAEVAEL
Mvan_0245|M.vanbaalenii_PYR-1 PLSDDEIRDELIVFMLAGHDTTATTLTYAMWALGHHHDVQDRVRAEVVGI
MUL_0604|M.ulcerans_Agy99 SLSDQQICDEMVYFMILGEDTTSTVLTYALWALGQHCQLQDRLAAEAAEL
MSMEG_0822|M.smegmatis_MC2_155 GLTDDQICDELVLFMLAGHDTTSTTLCYSLWALGHHPDIQERVYAEVAAL
Mb1429c|M.bovis_AF2122/97 IWPRQRVRDEALTFMLAGHETTANAMSWFWYLMALNPQARDHMLTELDDV
Rv1394c|M.tuberculosis_H37Rv IWPRQRVRDEALTFMLAGHETTANAMSWFWYLMALNPQARDHMLTELDDV
MMAR_0281|M.marinum_M AFTDTEVIDQIVSFFIAGTETTALTVAWALYLLAEHPAIEQRVHAEIDPV
MAV_3106|M.avium_104 ALNDNEIRDAIVSLVTAGYETTSGALAWAVYLLLSQPGAWATAAGEVRRV
MAB_2151c|M.abscessus_ATCC_199 QIRERRRRSEAAGLVFAGFETTAAALAWTIALLCRNPIALGKAYAEVDAL
MLBr_02088|M.leprae_Br4923 QLTEEEIIATCGLLLVAGHETTVNLIANAVLAMLRNPSQWKALSSNPQRA
:. * :** : : : :
Mflv_0056|M.gilvum_PYR-GCK G-RRELEPGDVPHLKYTTQVLLEALRLCPPAAGVGRMAVRDIDVDGHRVQ
TH_3255|M.thermoresistible__bu P-DREITPHDVPRLGYTVQVLRESLRLCPPAAGVGRLAMSDLAVDGYRVE
Mvan_0245|M.vanbaalenii_PYR-1 G-DRELTPEDVPALGYTIQVLHEALRLCPPAAATARMAMRDVEVDGYRIE
MUL_0604|M.ulcerans_Agy99 G-DRPLTPADVPRLGYTIQVLQEAMRLRPPGPTVARRAMQDIEVDGYRVQ
MSMEG_0822|M.smegmatis_MC2_155 G-DRELTTDDVPQLTYTMRVLHEALRLCPPGSGTMRMLNEEMTVDGYRVE
Mb1429c|M.bovis_AF2122/97 LGMRRPTADDLGKLAWTTACLQESQRYFSSVWIIAREAVDDDIIDGHRIR
Rv1394c|M.tuberculosis_H37Rv LGMRRPTADDLGKLAWTTACLQESQRYFSSVWIIAREAVDDDIIDGHRIR
MMAR_0281|M.marinum_M LAGRAATHADLPRLGLVNRIVTETLRLYPPGWIISRVATADVYLGGHRIP
MAV_3106|M.avium_104 LAGRLPAAADLSGLTYLNGVVHETLRLYPPGVISARRVMRDLRFEGRRIR
MAB_2151c|M.abscessus_ATCC_199 G-GKTLAYDDLENLKYLRACFDEAQRFQAAP-ANVRTAIEDDEVGGYFIP
MLBr_02088|M.leprae_Br4923 P-----------------LVVEETLRYDPAIHLIGRVAAKDMTIGQTTLT
. *: * .. * : . :
Mflv_0056|M.gilvum_PYR-GCK AGTLVAVGIYALHRDPEIWPDPDRFDPDRFSPEQTAQ--RDRWHFLPFAG
TH_3255|M.thermoresistible__bu AGTLLAVGIQAIHRDPSLWPDPMVFDPDRFAPERFKA--MDRWQFVPFIG
Mvan_0245|M.vanbaalenii_PYR-1 AGSMMAVGIYALHRDPALWERPLVFDPDRFSAQNSGG--RDRWQYLPFGA
MUL_0604|M.ulcerans_Agy99 AGTYCLVGVYAMHRDPALWDDPLAFDPDRFAPKKSKG--RDRWQYLPFGG
MSMEG_0822|M.smegmatis_MC2_155 AGTVAIVNFYAMHRSPTLWDAPERFDPDRFSPERSAG--RNRWQYLPFGG
Mb1429c|M.bovis_AF2122/97 RGTTVVIPIHHIHHDPRWWPDPDRFDPGRFLRCPTD---RPRCAYLPFGG
Rv1394c|M.tuberculosis_H37Rv RGTTVVIPIHHIHHDPRWWPDPDRFDPGRFLRCPTD---RPRCAYLPFGG
MMAR_0281|M.marinum_M AGATVIFSPYLIHHRPDLYAEPERFDPDRWLPERARA--IGRNTYIPFGG
MAV_3106|M.avium_104 SGRLLIFSPYVTHRLPEIWPEPMRFAPERWNPDAPGYRRPAPHEFIPFSA
MAB_2151c|M.abscessus_ATCC_199 RGSQVIITQYALQRDPRFWNEPERFNPDRFLTDRIN-----RNTFLPFSI
MLBr_02088|M.leprae_Br4923 EGDTMVLLLAAANRDPAVYSRPDEFDPDRPSS-----------RHLAFAV
* . :: * : * * * * .:.*
Mflv_0056|M.gilvum_PYR-GCK GARSCIGEHFARLETTLALATIVRSMRVTSSDNDFPVDVP-FTTVAHGPI
TH_3255|M.thermoresistible__bu GPRSCIGEHFAMLESTLAIATLIRSVRIRSLDDDFPVEVP-FTTVARGPI
Mvan_0245|M.vanbaalenii_PYR-1 GPRSCIGDHFAMLEATLALATIIRSAEIRSSDPHFPMIVP-FTTVAAEPI
MUL_0604|M.ulcerans_Agy99 GGRSCLGDHFAMLEASLALATIIREAEIHSLDGDFPVAIP-LPIVPAGPI
MSMEG_0822|M.smegmatis_MC2_155 GPRSCIGDHFAMLEATLALATVIRAVSVESQNTDLPVETP-FTVIAAAPV
Mb1429c|M.bovis_AF2122/97 GRRICIGQSFALMEMVLMAAIMSQHFTFDLAPG-YHVELE-ATLTLRPKH
Rv1394c|M.tuberculosis_H37Rv GRRICIGQSFALMEMVLMAAIMSQHFTFDLAPG-YHVELE-ATLTLRPKH
MMAR_0281|M.marinum_M GARKCIGDNFGTTEAALALATIAANWRMQRLPGLPVRPAV-AASLRPRNL
MAV_3106|M.avium_104 GLHRCVGAAMATTEMTVMLARLLARTRLRLPAQRLRAANV-AALRPTPGL
MAB_2151c|M.abscessus_ATCC_199 GPRKCMGTRMAYIEGTLVLGAILQRYAFQIRDGWTPRHRVRVSTGLAGGL
MLBr_02088|M.leprae_Br4923 GSHFCLGAALARLEATVTLSAISARFPQVQLAGELVYKPN-VAMRGMSAL
* : *:* :. * : . : .
Mflv_0056|M.gilvum_PYR-GCK PAHLTPR-------------------
TH_3255|M.thermoresistible__bu RAVVSARR------------------
Mvan_0245|M.vanbaalenii_PYR-1 RARVKSVNR------IPAADSGGR--
MUL_0604|M.ulcerans_Agy99 RARVNQRRPSQEVCTTTLSDSSASS-
MSMEG_0822|M.smegmatis_MC2_155 PARVTRR-------------------
Mb1429c|M.bovis_AF2122/97 GVHVIGRRR-----------------
Rv1394c|M.tuberculosis_H37Rv GVHVIGRRR-----------------
MMAR_0281|M.marinum_M ALRVTARTADRVPGEPGRRLTSREIG
MAV_3106|M.avium_104 TVEVIDSVPAQ---------------
MAB_2151c|M.abscessus_ATCC_199 PARLFAR-------------------
MLBr_02088|M.leprae_Br4923 PVQV----------------------
: