For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVDRASRLSIRAQIVQQLRSALESGVLGPGSPVPSSRAVASALGVSRSTVVAALQELEGEGWIESSQGSG TVVSHRPHATSDVARDPAGSATMSDEDIEIDMRPGDLNPGDISRVGWRAAWRSIEPSTTPPPPPGSAALR AALSTYLRSSRGLPCDPSQIVVCAGTAEALTLLTLAFQWRERAVAVEDPGYPDIRRTFQTLGVRWVPVDV TDADQFLSGLQSIEKPIEAAYLTPSHQYPLGHRISPGVRGDILRWARDAGVILVEDDYDSEFRFGVAPLP SLAGLDTTASTIYLGTMSKVLDPGLRLSYLRVPTHLLQPVLDTRDALGATVSAPVQQAVTHLLTSGELAR HIARVRKLYQDRRATMLQSLEKIAAVTHVTGTEAGLHVLASLRQGISASAAVEQARSHRYAHHRPRRLPG PSRPVESRAGPRIRTAQTGHNPARGQPTGHPARITGGRRPVTGDRAQIQTPLPTATTPPALTLKRSRSRS ISTPTCSPTPTTTFLSRIACRTTAPRPTRTPGISTLPSTTAPSSITTLEDSTELRTDAADTMLPAPTTDS CADPPLTNLAGARFSDSPRIGQRRLYRLNTGCTDTRSMCAS*
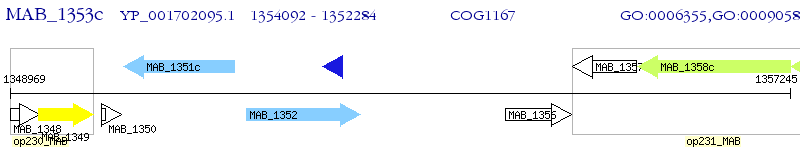
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1353c | - | - | 100% (602) | GntR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0196c | - | 5e-47 | 32.74% (391) | GntR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3582 | - | 3e-20 | 26.11% (360) | GntR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0881c | - | 7e-10 | 25.49% (255) | aminotransferase |
| M. gilvum PYR-GCK | Mflv_1181 | - | 6e-48 | 33.33% (393) | GntR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0858c | - | 7e-10 | 25.49% (255) | aminotransferase |
| M. leprae Br4923 | MLBr_01488 | - | 9e-07 | 26.77% (269) | N-succinyldiaminopimelate aminotransferase |
| M. marinum M | MMAR_2376 | - | 2e-21 | 27.32% (377) | GntR family transcriptional regulator |
| M. avium 104 | MAV_3214 | - | 3e-19 | 27.47% (375) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5760 | - | 4e-57 | 36.50% (400) | GntR-family protein regulator |
| M. thermoresistible (build 8) | TH_1831 | - | 1e-09 | 26.33% (281) | PROBABLE AMINOTRANSFERASE |
| M. ulcerans Agy99 | MUL_1552 | - | 8e-22 | 26.67% (435) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5625 | - | 6e-45 | 32.06% (393) | GntR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1181|M.gilvum_PYR-GCK MVSSWSNSGSRDLHLDLREALRPGARGNRNALVAALREAVRSGRLSAGTT
Mvan_5625|M.vanbaalenii_PYR-1 -MTSWANSATRDLHLDLRDALVPGARGTKDALISALRDAARSGRLTAGTM
MSMEG_5760|M.smegmatis_MC2_155 ----MDRIAGLDLHLDR-----PAGR-TREALMDAVRDAIRSGRLVSGTR
MMAR_2376|M.marinum_M -MPVQYRIAG----------------GGAESIAASIESAISQGGLAPGDA
MUL_1552|M.ulcerans_Agy99 -MPVQNRIAG----------------GGAGSIAASIESAISQGGLAPGDA
MAV_3214|M.avium_104 -MAVQKSITG----------------TGAESIAASIEAAISAGSLAPGDA
Mb0881c|M.bovis_AF2122/97 ---------------------------------------MTVSRLRPYAT
Rv0858c|M.tuberculosis_H37Rv ---------------------------------------MTVSRLRPYAT
MLBr_01488|M.leprae_Br4923 --------------------------------MSPFGRVSATLPVFPWDT
TH_1831|M.thermoresistible__bu --------------------------------------------------
MAB_1353c|M.abscessus_ATCC_199 --MRVDRASR---------------LSIRAQIVQQLRSALESGVLGPGSP
Mflv_1181|M.gilvum_PYR-GCK LPPSRALAADLGLARNTVADAYAELVAEGWLASRQGAGTWVVDTARASSQ
Mvan_5625|M.vanbaalenii_PYR-1 LPSSRTLATDLGLARNTVAEAYAELVAEGWLASRQGAGTWVADVT----T
MSMEG_5760|M.smegmatis_MC2_155 LPSSRALAADLGVARNTVARAYAELIAEGWLTSQHGSHTTVSQRAAEAVR
MMAR_2376|M.marinum_M LPSIREVAGQLGVNPNTVAAAYRLLRDRGTVETAGRRGTRVRDRPASTPR
MUL_1552|M.ulcerans_Agy99 LPSIREVAGQLGVNPNTVAAAYRLLRDRGTVETAGRRGTRVRDRPASTPR
MAV_3214|M.avium_104 LPPVRELAARLGVNANTAAAAYRLLRHRGAVETAGRRGTRVRHRPATTPR
Mb0881c|M.bovis_AF2122/97 TVFAEMSALATRIGAVNLGQGFPDEDGPPKMLQAAQDAIAG---------
Rv0858c|M.tuberculosis_H37Rv TVFAEMSALATRIGAVNLGQGFPDEDGPPKMLQAAQDAIAG---------
MLBr_01488|M.leprae_Br4923 LADAKAVAESHPDGIVDLSVGTPVDPVAPLIQEALAAASS----------
TH_1831|M.thermoresistible__bu --------------------------VAPVIRAALAEASA----------
MAB_1353c|M.abscessus_ATCC_199 VPSSRAVASALGVSRSTVVAALQELEGEGWIESSQGSGTVVSHRPHATSD
:
Mflv_1181|M.gilvum_PYR-GCK PPPVPPRPHGARV-APVHNLMPGSPDVAEFPRSQWAASARRALTHAPTDA
Mvan_5625|M.vanbaalenii_PYR-1 PAHTPPRPHGVRV-APTHNLMPGSPDVAEFPRSQWLASTRRALGGAPTEA
MSMEG_5760|M.smegmatis_MC2_155 SVTPPARPRPPR--RLDHDLRPGHPDLSSFPRTEWSRAVKRALANAPSEA
MMAR_2376|M.marinum_M SSLDLGVPAGAR------DLSTGNPAPELLP---------LAAVG-WPDG
MUL_1552|M.ulcerans_Agy99 SSLDLGVPAGAR------DLSTGNPAPELLP---------LAAVG-WPDG
MAV_3214|M.avium_104 SLLGLDVPAGVR------DLSTGNPHPALLP---------LAAAAPARPA
Mb0881c|M.bovis_AF2122/97 ------------------------------------------------GV
Rv0858c|M.tuberculosis_H37Rv ------------------------------------------------GV
MLBr_01488|M.leprae_Br4923 -------------------------------------------------V
TH_1831|M.thermoresistible__bu -------------------------------------------------A
MAB_1353c|M.abscessus_ATCC_199 VARDPAGSATMSDEDIEIDMRPGDLNPGDISRVG-----WRAAWRSIEPS
Mflv_1181|M.gilvum_PYR-GCK FRMGDPRGRPELRSALAEYLARARGVR-TSPESLVICAGVRHAVELLART
Mvan_5625|M.vanbaalenii_PYR-1 LRMGDPRGRPELRSALAEYLARARGVR-TSPESIVICAGVRHAVELLTRA
MSMEG_5760|M.smegmatis_MC2_155 FGYAEPQGRPELRRALAEYLARARGVR-ARPCNIVVCCGAAEGLNLVAGA
MMAR_2376|M.marinum_M SVLPLLYGAPAMSAELVDYARAALAADGVAADHLAVTNGALDGIERALSA
MUL_1552|M.ulcerans_Agy99 SVLPLLYGAPAMSAELVDYARAALAADGVAADHLAVTNGALDGIERALSA
MAV_3214|M.avium_104 SGRAVLYGEPAISPALAEYARAALSADGVPADHLALTSGALDGIERALTA
Mb0881c|M.bovis_AF2122/97 NQYPPGPGSAPLRRAIAAQRRRHFGVDYDPETEVLVTVGATEAIAAAVLG
Rv0858c|M.tuberculosis_H37Rv NQYPPGPGSAPLRRAIAAQRRRHFGVDYDPETEVLVTVGATEAIAAAVLG
MLBr_01488|M.leprae_Br4923 AGYPTTAGTARLREAAVAALDRRYGITGLTEAAVLPVIGTKELIAWLPTL
TH_1831|M.thermoresistible__bu PGYPTTAGTPALRASAAAALERRYGITGLTPDAVLPAIGTKELIAWLPTL
MAB_1353c|M.abscessus_ATCC_199 TTPPPPPGSAALRAALSTYLRSSRGLP-CDPSQIVVCAGTAEALTLLTLA
* . : . : *. . :
Mflv_1181|M.gilvum_PYR-GCK LG----GPVAVEAYGLFVFRDAIAACGVQTVPIGVDDDG---AVIADLDD
Mvan_5625|M.vanbaalenii_PYR-1 LG----GPIAVESYGLFVFRDAIAAAGASTVPIGVDDRG---AVVDELDG
MSMEG_5760|M.smegmatis_MC2_155 LAEMGVAATAVEAYGLPTQRAALARAGLRCPPLAVDADG---ADINALNT
MMAR_2376|M.marinum_M NLRPG-DRVAVEDPGWANLLDLLAALGLPVEPMGVDDDG---PLVADLQR
MUL_1552|M.ulcerans_Agy99 NLRPG-DRVAVEDPGWANLLDLLAALGLPVEPMGVDDDG---PLVADLQR
MAV_3214|M.avium_104 HLRPG-DRVAVEDPGWANLLDLLAALGFSAEPVRVDDAG---PLPEDLAR
Mb0881c|M.bovis_AF2122/97 L-VEPGSEVLLIEPFYDSYSPVVAMAGAHRVTVPLVPDGRGFALDADALR
Rv0858c|M.tuberculosis_H37Rv L-VEPGSEVLLIEPFYDSYSPVVAMAGAHRVTVPLVPDGRGFALDADALR
MLBr_01488|M.leprae_Br4923 LGLRAGDVVVVPELAYPTYNVGARLAGAR-------------VLRADSLT
TH_1831|M.thermoresistible__bu LGLGGDDTVVVPELAYPTYDVGARLAGAR-------------VFRADSLT
MAB_1353c|M.abscessus_ATCC_199 FQWR-ERAVAVEDPGYPDIRRTFQTLGVRWVPVDVTDAD----QFLSGLQ
: *
Mflv_1181|M.gilvum_PYR-GCK T---SARSVLLTPAHHNPLGMPLHPSRRAAVIEWATRTGGYILDDDYDGE
Mvan_5625|M.vanbaalenii_PYR-1 L---DVPAVLLTPAHHNPLGMPLHSSRRTAVIEWAQRTGGFVLDDDYDGE
MSMEG_5760|M.smegmatis_MC2_155 MP--EVGAVLLTPSHQFPLGVPLHSDRRAAVIDWARRTGGMIIEDDYDGE
MMAR_2376|M.marinum_M ALGRGVRAVVVTSRAQNPTGAALSAARAEELREVLSQGAEEVLLVEDDHC
MUL_1552|M.ulcerans_Agy99 ALGRGVRAVVVTSRAQNPTGAALSAARAEELREVLSQGAEEVLLVEDDHC
MAV_3214|M.avium_104 ALGRGARALVLTTRAQNPTGAALSAERAAALRDLLSGRADDVLLVEDDHC
Mb0881c|M.bovis_AF2122/97 RAVTPRTRALIINSPHNPTGAVLSATELAAIAEIAVAANLVVITDEVYEH
Rv0858c|M.tuberculosis_H37Rv RAVTPRTRALIINSPHNPTGAVLSATELAAIAEIAVAANLVVITDEVYEH
MLBr_01488|M.leprae_Br4923 QLGLQSPALFYLNSPSNPTGQVLAVNHLRKMVGWARNRGVVVVSDECYLG
TH_1831|M.thermoresistible__bu QLGPQTPALIYLNSPSNPTGRVLGVDHLRKVVGWARDRGVIVASDECYLG
MAB_1353c|M.abscessus_ATCC_199 SIEKPIEAAYLTPSHQYPLGHRISPGVRGDILRWARDAGVILVEDDYDSE
* * : : : :
Mflv_1181|M.gilvum_PYR-GCK FRYDRQPVGALQALRP-DRVAYLGSTSKSLS-----QALRLGWMALPAEL
Mvan_5625|M.vanbaalenii_PYR-1 FRYDRQPVGALQSLRP-DRVAYLGSTSKSLS-----QTLRLGWMALPDNL
MSMEG_5760|M.smegmatis_MC2_155 FRYDRSPVGALHGVDP-AHVIYLGTASKSLA-----PGLRLGWLVLPDRL
MMAR_2376|M.marinum_M AGIAGVPLHTLAGAT--QHWAFVRSAAKAYG-----PDLRLAVLAGDQRT
MUL_1552|M.ulcerans_Agy99 AGIAGVPLHTLAGAT--QHWAFVRSAAKAYG-----PDLRLAVLAGDQRT
MAV_3214|M.avium_104 AGISGAALHPVAGCT--THWAFVRSASKAYG-----PDLRVAVLAGDQRT
Mb0881c|M.bovis_AF2122/97 LVFDHARHLPLAG--F-DGMAERTITISSAAKMFNCTGWKIGWACGPAEL
Rv0858c|M.tuberculosis_H37Rv LVFDHARHLPLAG--F-DGMAERTITISSAAKMFNCTGWKIGWACGPAEL
MLBr_01488|M.leprae_Br4923 LGWDAEPFSVLHPLVC-DGNHTGLLAVHSLSKSSSLAGYRAGFVAGDPDL
TH_1831|M.thermoresistible__bu LGWDETPVSVLHPTVC-GGDHTNLLAIHSLSKTSSLAGYRAGFVAGDPAL
MAB_1353c|M.abscessus_ATCC_199 FRFGVAPLPSLAGLDTTASTIYLGTMSKVLD-----PGLRLSYLRVPTHL
: : .
Mflv_1181|M.gilvum_PYR-GCK VDQVVETAGGQQFHVDVVSQLTLADFLTAGHYDRHIRRMRMRYRRRRDML
Mvan_5625|M.vanbaalenii_PYR-1 IDAVLAAAGGQQFYVDAINQLTLADFIAGGHYDRHIRRMRMRYRRRRDVL
MSMEG_5760|M.smegmatis_MC2_155 VDAVLRQKGETEETSGFVDQLTMAEFIESGSYDRHIRTMRAQYRRRREQL
MMAR_2376|M.marinum_M VERVHGRLRLGPGWVSHILQDLAVRLFADDAATGLVRQAQERYADARGRL
MUL_1552|M.ulcerans_Agy99 VERVHGRLRLGPGWVSHILQDLAVRLFADDAATGLVRQAQERYADARGRL
MAV_3214|M.avium_104 VDRVHGRLRLGPGWVSHVLQQLAVELWSDTAASELVATAERRYADRRRRL
Mb0881c|M.bovis_AF2122/97 IAGVRAAKQYLSYVGGAPFQPAVALALDT--EDAWVAALRNSLRARRDRL
Rv0858c|M.tuberculosis_H37Rv IAGVRAAKQYLSYVGGAPFQPAVALALDT--EDAWVAALRNSLRARRDRL
MLBr_01488|M.leprae_Br4923 VAELLAVRKHAGMMVPTPVQAAMVAALG---DDAHEREQRDRYARRRDVL
TH_1831|M.thermoresistible__bu VAELLAVRKHAGMMVPTPVQAAMVAALD---DDAHERQQRERYQRRRREL
MAB_1353c|M.abscessus_ATCC_199 LQPVLDTRDALGATVSAPVQQAVTHLLTSGELARHIARVRKLYQDRRATM
: : * . . * :
Mflv_1181|M.gilvum_PYR-GCK VSALSPFAVG--IGGLAAGVNLLVTLPDGAEPELIRRAGEA---------
Mvan_5625|M.vanbaalenii_PYR-1 VSALSPFDVG--IGGLAAGVNLLVTLPDGAEPEVMRRAGES---------
MSMEG_5760|M.smegmatis_MC2_155 VAAVTAASPATTVAGLPAGLHVMLEIAGANASALAHQLAWR---------
MMAR_2376|M.marinum_M CAALSARGVA---AHGRSGLNVWIPVPD--ETAAITRLLNR---------
MUL_1552|M.ulcerans_Agy99 CAALSARGVA---AHGRSGLNVWIPVPD--ETAAITRLLNR---------
MAV_3214|M.avium_104 CGALAERGVD---AHGRSGLNVWVRVPD--EAVAVSRLLGA---------
Mb0881c|M.bovis_AF2122/97 AAGLTEIGFA--VHDSYGTYFLCADPRPLGYDDSTEFCAALP--------
Rv0858c|M.tuberculosis_H37Rv AAGLTEIGFA--VHDSYGTYFLCADPRPLGYDDSTEFCAALP--------
MLBr_01488|M.leprae_Br4923 LPAVVSAGFT--VDHSEAGLYLWAS-RGEPCRDSVEWFAQR---------
TH_1831|M.thermoresistible__bu LPALQQAGFT--VDHSEAGLYLWAT-RGEPCRATVDWLARR---------
MAB_1353c|M.abscessus_ATCC_199 LQSLEKIAAVTHVTGTEAGLHVLASLRQGISASAAVEQARSHRYAHHRPR
.: . :
Mflv_1181|M.gilvum_PYR-GCK -------GIALVGLSMMRHPRAGSG-----------------------VA
Mvan_5625|M.vanbaalenii_PYR-1 -------GLAVSGLSIMRHPMAGP-------------------------A
MSMEG_5760|M.smegmatis_MC2_155 -------RLGVESLDRYRHPEFDN--------------------------
MMAR_2376|M.marinum_M -------GWAAAPGARFRIRSAPGI-----------------------RV
MUL_1552|M.ulcerans_Agy99 -------GWAAAPGARFRVRSAPGI-----------------------RV
MAV_3214|M.avium_104 -------GWAAAPGARFRMQTPAGI-----------------------RI
Mb0881c|M.bovis_AF2122/97 ----EKVGVAAIPMSAFCDPAAGQASQQ--------------------AD
Rv0858c|M.tuberculosis_H37Rv ----EKVGVAAIPMSAFCDPAAGQASQQ--------------------AD
MLBr_01488|M.leprae_Br4923 --------GILVALGEFYGPGG----------------------------
TH_1831|M.thermoresistible__bu --------GILVAPGEFYGPRG----------------------------
MAB_1353c|M.abscessus_ATCC_199 RLPGPSRPVESRAGPRIRTAQTGHNPARGQPTGHPARITGGRRPVTGDRA
Mflv_1181|M.gilvum_PYR-GCK ERDGLIVGFAAAPEHAFPAAVSALCEVLRASGL-----------------
Mvan_5625|M.vanbaalenii_PYR-1 DVDGLIVGFAAPPEHAFSAAVAALCDVLRASGL-----------------
MSMEG_5760|M.smegmatis_MC2_155 ERSGLVFSYAAPSPSAWSAALDAVVRLLP---------------------
MMAR_2376|M.marinum_M TIAGLSAEEIEPLADAIAEAIQRTGRPTV---------------------
MUL_1552|M.ulcerans_Agy99 TIAGLSAEEIEPLADAIAGAIQRTGRPTV---------------------
MAV_3214|M.avium_104 TIADLTPDEIEPLADAVAQAVRPSGRPIV---------------------
Mb0881c|M.bovis_AF2122/97 VWNHLVRFTFCKRDDTLDEAIRRLSVLAERPAT-----------------
Rv0858c|M.tuberculosis_H37Rv VWNHLVRFTFCKRDDTLDEAIRRLSVLAERPAT-----------------
MLBr_01488|M.leprae_Br4923 --CQHVRVALTASDERIAAVVARLS-------------------------
TH_1831|M.thermoresistible__bu --AEHVRIALTAPDERITAAAARLAQAG----------------------
MAB_1353c|M.abscessus_ATCC_199 QIQTPLPTATTPPALTLKRSRSRSISTPTCSPTPTTTFLSRIACRTTAPR
Mflv_1181|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5625|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5760|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2376|M.marinum_M --------------------------------------------------
MUL_1552|M.ulcerans_Agy99 --------------------------------------------------
MAV_3214|M.avium_104 --------------------------------------------------
Mb0881c|M.bovis_AF2122/97 --------------------------------------------------
Rv0858c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01488|M.leprae_Br4923 --------------------------------------------------
TH_1831|M.thermoresistible__bu --------------------------------------------------
MAB_1353c|M.abscessus_ATCC_199 PTRTPGISTLPSTTAPSSITTLEDSTELRTDAADTMLPAPTTDSCADPPL
Mflv_1181|M.gilvum_PYR-GCK -----------------------------------
Mvan_5625|M.vanbaalenii_PYR-1 -----------------------------------
MSMEG_5760|M.smegmatis_MC2_155 -----------------------------------
MMAR_2376|M.marinum_M -----------------------------------
MUL_1552|M.ulcerans_Agy99 -----------------------------------
MAV_3214|M.avium_104 -----------------------------------
Mb0881c|M.bovis_AF2122/97 -----------------------------------
Rv0858c|M.tuberculosis_H37Rv -----------------------------------
MLBr_01488|M.leprae_Br4923 -----------------------------------
TH_1831|M.thermoresistible__bu -----------------------------------
MAB_1353c|M.abscessus_ATCC_199 TNLAGARFSDSPRIGQRRLYRLNTGCTDTRSMCAS