For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVLPLLPRGELLASLSDEMLARRAQQGDSEAFDVLVVRHGPALLRFVARTYPERAEYDDIVQETFIAAWK GLPSFAFRSQFRTWLYSLASRKTVDAMRRRRRDFDIDEVPEVADAGAGPSERALDGDFLAALSRELSQLP YAARAAWWLREVYDLPLAEIATVLRTTEGSVRGNLQRTRKRLAETLKDFRP
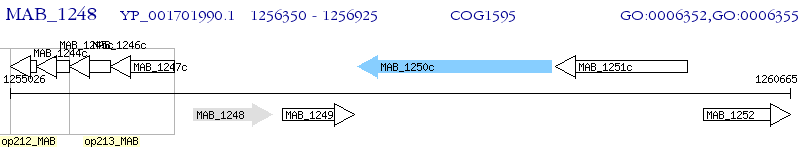
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1248 | - | - | 100% (191) | RNA polymerase sigma factor |
| M. abscessus ATCC 19977 | MAB_3543c | - | 7e-12 | 26.70% (191) | RNA polymerase sigma-E factor |
| M. abscessus ATCC 19977 | MAB_3428c | - | 1e-11 | 33.54% (164) | RNA polymerase sigma-C factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3448c | sigD | 3e-13 | 30.98% (184) | RNA polymerase sigma factor SigD |
| M. gilvum PYR-GCK | Mflv_3908 | - | 7e-13 | 32.70% (159) | ECF subfamily RNA polymerase sigma-24 factor |
| M. tuberculosis H37Rv | Rv3414c | sigD | 3e-13 | 30.98% (184) | RNA polymerase sigma factor SigD |
| M. leprae Br4923 | MLBr_01448 | - | 2e-10 | 29.03% (186) | RNA polymerase sigma factor SigC |
| M. marinum M | MMAR_1134 | sigD | 1e-13 | 31.69% (183) | alternative RNA polymerase sigma-D factor, SigD |
| M. avium 104 | MAV_4359 | - | 3e-12 | 28.80% (184) | RNA polymerase sigma factor SigD |
| M. smegmatis MC2 155 | MSMEG_6931 | - | 2e-15 | 30.37% (191) | RNA polymerase sigma-70 factor |
| M. thermoresistible (build 8) | TH_0604 | sigG | 2e-12 | 32.20% (177) | PROBABLE ALTERNATIVE RNA POLYMERASE SIGMA FACTOR SIGG (RNA |
| M. ulcerans Agy99 | MUL_0897 | sigD | 9e-14 | 31.69% (183) | RNA polymerase sigma factor SigD |
| M. vanbaalenii PYR-1 | Mvan_0782 | - | 2e-15 | 32.52% (206) | ECF subfamily RNA polymerase sigma-24 factor |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3448c|M.bovis_AF2122/97 MVDPGVSPGCVRFVTLEISPSMTMQGERLDAVVAEAVAGDRNALREVLET
Rv3414c|M.tuberculosis_H37Rv MVDPGVSPGCVRFVTLEISPSMTMQGERLDAVVAEAVAGDRNALREVLET
MMAR_1134|M.marinum_M ---------------MEISPSMTIEGERLDAVVAEAVAGDRNALREVLET
MUL_0897|M.ulcerans_Agy99 ---------------MEISPSMTIEGERLDAVVAEAVAGDRNALREVLET
MAV_4359|M.avium_104 ---------------MTIP-----AGERLDAVVAKAVTGDHNALREVLET
Mflv_3908|M.gilvum_PYR-GCK -------------------------MISHPGVPRRLHDVKVRPFEEIVAC
MAB_1248|M.abscessus_ATCC_1997 ------------MVLPLLPRGELLASLSDEMLARRAQQGDSEAFDVLVVR
TH_0604|M.thermoresistible__bu -------------------MSVMLRTLLHVTVLTAPPDGGSDAFLADAEQ
MSMEG_6931|M.smegmatis_MC2_155 --------------MGRGEFAGAAHTRSDAELLAAHVAGDRYAFEELVLR
MLBr_01448|M.leprae_Br4923 -------------------MIASSGDEAVTDLALAAARGSKRALEAFIKA
Mvan_0782|M.vanbaalenii_PYR-1 -------------MTGYHLAEMTTTSADEADLIAALRARDEVAFARLVDQ
: .:
Mb3448c|M.bovis_AF2122/97 IRPIVVRYCRARVGTVERSGLSADDVAQEVCLATITALPRYRDRGRPFLA
Rv3414c|M.tuberculosis_H37Rv IRPIVVRYCRARVGTVERSGLSADDVAQEVCLATITALPRYRDRGRPFLA
MMAR_1134|M.marinum_M IRPIVVRYCRARVGTVERSGLSADDVAQEVCLATITALPRYRDRGRPFLA
MUL_0897|M.ulcerans_Agy99 IRPIVVRYCRARVGTVERSGLSADDVAQEVCLATITALPRYRDRGRPFLA
MAV_4359|M.avium_104 IRPIVVRYCRARVGTVERGGLSADDVAQEVCLATITALPRYRDRGRPFLA
Mflv_3908|M.gilvum_PYR-GCK HGATVLRVCRAVVGPVD-----AEDAWSETFLSALRAYPELPEDAN-VEA
MAB_1248|M.abscessus_ATCC_1997 HGPALLRFVARTYPERA----EYDDIVQETFIAAWKGLPSFAFRSQ-FRT
TH_0604|M.thermoresistible__bu YRRELLAHCYRMTGSLH----DAEDLVQETFLRAWKSHHKFEGKAS-VRT
MSMEG_6931|M.smegmatis_MC2_155 HHRQLRRLAYLTSNHLD----DADDAVQEALLKAHRTAATFRFDCA-VSS
MLBr_01448|M.leprae_Br4923 TQQDVWRFVAYLSDVGN-----ADDLTQETFLRAIGAIERFSGHSS-GRT
Mvan_0782|M.vanbaalenii_PYR-1 HTPAMLRVARGYVASHE----VAEEVVQETWIALLKGIDRFEGRSS-LRT
: :: .*. : :
Mb3448c|M.bovis_AF2122/97 FLYGIAAHKVADAHRAAGRD------RAYPAETLPERWSADAGPEQMAIE
Rv3414c|M.tuberculosis_H37Rv FLYGIAAHKVADAHRAAGRD------RAYPAETLPERWSADAGPEQMAIE
MMAR_1134|M.marinum_M FLYGIAAHKVADAHRAAGRD------LAYPAESVPERRSADAGPEQLAIE
MUL_0897|M.ulcerans_Agy99 FLYGIAAHKVADAHRAAGRD------LAYPAESVPERRSADAGPEQLAIE
MAV_4359|M.avium_104 FLYGIAAHKVADAHRAAGRD------LAYPTESIPDRWSNDAGPEQLAIE
Mflv_3908|M.gilvum_PYR-GCK WLVTIAHRRALDLGRARSR-------RAVPTDSVPERCDARADP------
MAB_1248|M.abscessus_ATCC_1997 WLYSLASRKTVDAMRRRRR--------DFDIDEVPEVADAGAGPSERALD
TH_0604|M.thermoresistible__bu WLYKIATNACLTALDGRQRRPLPTGLGAPSSDPTDDIVAREEVPWLEPLP
MSMEG_6931|M.smegmatis_MC2_155 WLYRIVVNACLDRLRRTRTQP--SATHYVELGHVNHLRDPAADPAPRVTT
MLBr_01448|M.leprae_Br4923 WLLAIARRVVADHIRRAQTR------PRTTFDGYPERMLNADRHARGFED
Mvan_0782|M.vanbaalenii_PYR-1 WLFTVMINIAKTRGLRDRTDADVQIAAYTGGTVDPKRFRDSDDPWPGHWK
:* : . .
Mb3448c|M.bovis_AF2122/97 ADSVT------------------RMNELLEILPAKQREILILRVVVGLSA
Rv3414c|M.tuberculosis_H37Rv ADSVT------------------RMNELLEILPAKQREILILRVVVGLSA
MMAR_1134|M.marinum_M ADSVT------------------RMNELLEILPAKQREILILRVVVGLSA
MUL_0897|M.ulcerans_Agy99 ADSVT------------------RMNELLEILPAKQREILILRVVVGLSA
MAV_4359|M.avium_104 ADSVS------------------RMSELLEILPAKQREILILRVVVGLSA
Mflv_3908|M.gilvum_PYR-GCK AARDP------------------DLWAALARLPDKQRHAVAYHHIAGLPF
MAB_1248|M.abscessus_ATCC_1997 GDFLA------------------ALSRELSQLPYAARAAWWLREVYDLPL
TH_0604|M.thermoresistible__bu DDAADPSTIVGSRESVR-----LAFVAALQHLPPRQRAVLLLREVLQWRA
MSMEG_6931|M.smegmatis_MC2_155 AITVE---------------------RALLQLPVDQRAAVVAVDMQGYSV
MLBr_01448|M.leprae_Br4923 LVEVT---------------------TMIANLATEQREALLLTQLLGLPY
Mvan_0782|M.vanbaalenii_PYR-1 ADEVPSPFPDTPEGSVLGGELLAVTRRELAKLPERQRLVVTMRDVLDMDS
: *. * :
Mb3448c|M.bovis_AF2122/97 EETAAAVGSTTGAVRVAQHRALQRLKDEIVAAGDYA--------------
Rv3414c|M.tuberculosis_H37Rv EETAAAVGSTTGAVRVAQHRALQRLKDEIVAAGDYA--------------
MMAR_1134|M.marinum_M EETAAAVGSTTGAVRVAQHRALQRLKDEIVAAGDYA--------------
MUL_0897|M.ulcerans_Agy99 EETAAAVGSTTGAVRVAQHRALQRLKDEIVAAGDYA--------------
MAV_4359|M.avium_104 EETAAAVGSTTGAVRVAQHRALSRLKSEMIAAGDCA--------------
Mflv_3908|M.gilvum_PYR-GCK TEIAVILGNSPEAARRAAADGIKALRSYYREDESA---------------
MAB_1248|M.abscessus_ATCC_1997 AEIATVLRTTEGSVRGNLQRTRKRLAETLKDFRP----------------
TH_0604|M.thermoresistible__bu AEVAEVLNTSTAAVNSLLQRARAQLDQVGPSEDDELVPPESPEAQELLTR
MSMEG_6931|M.smegmatis_MC2_155 AETARLLGVAEGTIKSRCARARAKLARTLQYFDGDEPQPATSAHRGAAHP
MLBr_01448|M.leprae_Br4923 ADAAAVCGCPVGTIRSRVARARDALLADTEHNHLTG--------------
Mvan_0782|M.vanbaalenii_PYR-1 AEVSRLLDLSPANQRVLLHRGRAAVRQALENYLKEPV-------------
: : . . :
Mb3448c|M.bovis_AF2122/97 --------------------------------------------------
Rv3414c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1134|M.marinum_M --------------------------------------------------
MUL_0897|M.ulcerans_Agy99 --------------------------------------------------
MAV_4359|M.avium_104 --------------------------------------------------
Mflv_3908|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1248|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0604|M.thermoresistible__bu YIDAFESYDIDKLVELFTADAVWEMPPFDGWYQGPQNIVTLSKTHCPAER
MSMEG_6931|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01448|M.leprae_Br4923 --------------------------------------------------
Mvan_0782|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3448c|M.bovis_AF2122/97 --------------------------------------------------
Rv3414c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1134|M.marinum_M --------------------------------------------------
MUL_0897|M.ulcerans_Agy99 --------------------------------------------------
MAV_4359|M.avium_104 --------------------------------------------------
Mflv_3908|M.gilvum_PYR-GCK --------------------------------------------------
MAB_1248|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0604|M.thermoresistible__bu PGDMRLIPTTANGQPAGAMYLRNPETGTHEAFQMHVLAIGPGGISHVVAF
MSMEG_6931|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01448|M.leprae_Br4923 --------------------------------------------------
Mvan_0782|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3448c|M.bovis_AF2122/97 --------------
Rv3414c|M.tuberculosis_H37Rv --------------
MMAR_1134|M.marinum_M --------------
MUL_0897|M.ulcerans_Agy99 --------------
MAV_4359|M.avium_104 --------------
Mflv_3908|M.gilvum_PYR-GCK --------------
MAB_1248|M.abscessus_ATCC_1997 --------------
TH_0604|M.thermoresistible__bu HMPSFAPFGLPDSL
MSMEG_6931|M.smegmatis_MC2_155 --------------
MLBr_01448|M.leprae_Br4923 --------------
Mvan_0782|M.vanbaalenii_PYR-1 --------------