For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARTDDDSWDLATGVGATATLVAAGRARAARAAQPLIDDPFAEPLVRAVGVEFLTRWATGELDAADVDDPD AAWGLQRMTTELVVRTRYFDQFFLDAAAAGVRQAVILASGLDARGYRLPWPADTTVFEVDQPRVLEFKAQ TLAGLGAQPTADLRMVPADLRHDWPDALRRGGFDAAEPAAWIAEGLFGYLPPDAQNRLLDHVTDLSAPGS RLALEAFLGSADRDSARVEEMIRTATRGWREHGFHLDIWALNYAGPRHEVSGYLDNHGWRSVGTTTAQLL AAHDLPAAPALPAGLADRPNYWTCVLG*
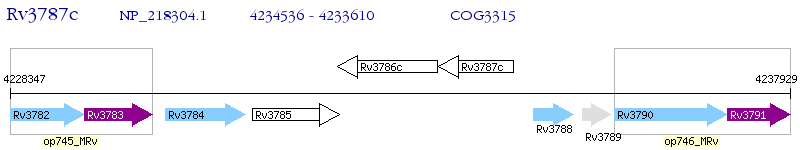
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3787c | - | - | 100% (308) | hypothetical protein Rv3787c |
| M. tuberculosis H37Rv | Rv0726c | - | 1e-85 | 55.83% (283) | hypothetical protein Rv0726c |
| M. tuberculosis H37Rv | Rv0731c | - | 1e-83 | 54.90% (286) | hypothetical protein Rv0731c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3816c | - | 0.0 | 100.00% (308) | hypothetical protein Mb3816c |
| M. gilvum PYR-GCK | Mflv_5023 | - | 4e-87 | 55.94% (286) | putative methyltransferase |
| M. leprae Br4923 | MLBr_02640 | - | 9e-55 | 44.36% (275) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_3787 | - | 6e-72 | 49.33% (300) | hypothetical protein MAB_3787 |
| M. marinum M | MMAR_3534 | - | 1e-100 | 63.89% (288) | O-methyltransferase |
| M. avium 104 | MAV_2110 | - | 1e-101 | 62.85% (288) | methyltransferase, putative, family protein |
| M. smegmatis MC2 155 | MSMEG_1888 | - | 2e-92 | 58.69% (305) | methyltransferase |
| M. thermoresistible (build 8) | TH_0301 | - | 4e-80 | 57.47% (261) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2766 | - | 1e-100 | 63.89% (288) | O-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1346 | - | 7e-86 | 54.64% (291) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3787c|M.tuberculosis_H37Rv MARTDDDSWDLATGVGATATLVAAGRARAARAAQPLIDDPFAEPLVRAVG
Mb3816c|M.bovis_AF2122/97 MARTDDDSWDLATGVGATATLVAAGRARAARAAQPLIDDPFAEPLVRAVG
MSMEG_1888|M.smegmatis_MC2_155 MEST-HETWDLATSVGATATMVAAGRARATTTG--LIDDRFAEPLVRAVG
MMAR_3534|M.marinum_M MPRTDNDSWDLATSVGATATMVAAARAIATNADNPLIADRFAEPLVRAVG
MUL_2766|M.ulcerans_Agy99 MPRTDNDSWDLATSVGATATMVAAARAIATNADNPLIADRFAEPLVRAVG
MAV_2110|M.avium_104 MPRTDNDTWDLSTSVGATATMVAAARAIATNADNPLIEDRFAEPLVRAVG
Mflv_5023|M.gilvum_PYR-GCK MPRTENDTWDLASSVGATATMVAAARAVATRAEDPVIDDPYAEPLVRAVG
Mvan_1346|M.vanbaalenii_PYR-1 MARTDNDTWDLASSVGATATMVAAARAVATRAPRPVIDDPFAAPLVQAVG
TH_0301|M.thermoresistible__bu MX-------------GG-----------ASRDPKELINDPFAAPLVRAVG
MAB_3787|M.abscessus_ATCC_1997 MARTDDDSWDLASSVGATATMVAAQRALASHVPNPLINDPYAEPLVRAVG
MLBr_02640|M.leprae_Br4923 -MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLLVTNTG
* : :* * :* ** .*
Rv3787c|M.tuberculosis_H37Rv VEFLTRWATGELDAADVDDP--DAAWGLQRMTTELVVRTRYFDQFFLDAA
Mb3816c|M.bovis_AF2122/97 VEFLTRWATGELDAADVDDP--DAAWGLQRMTTELVVRTRYFDQFFLDAA
MSMEG_1888|M.smegmatis_MC2_155 IDFMTRWATGDLAAADVDIP--GATWGMQQMTDLLAARTRYFDAFFGDAA
MMAR_3534|M.marinum_M VDFFTRWVTGDLVVADVDDT--ESGWQLAQMPDAMAVRTRFFDAFFQDAT
MUL_2766|M.ulcerans_Agy99 VDFFTRWVTGDLVVADVDDT--ESGWQLAQMPDAMAVRARFFDAFFQDAT
MAV_2110|M.avium_104 VDFFTRWVTGDLVAADVDDH--DSGWKLEHMPVAMAARTRFFDSFFQGAT
Mflv_5023|M.gilvum_PYR-GCK IDFFAKLASGELTPEDLDVSGDESPVGMSRFADGMAARTRFFDDFFTDAC
Mvan_1346|M.vanbaalenii_PYR-1 VDFFTRLATGELTTEELDTNDAESPVGMSRFADAMAARTRFFDDFFAEAM
TH_0301|M.thermoresistible__bu IDFFTRIADGELTVADLDP---QSAPRVQANIDEVAVRTRFFDDHFAAAT
MAB_3787|M.abscessus_ATCC_1997 IEYFTKLASGAFDPEQMAGL-VQLGITADGMANGMASRTWYYDTAFESST
MLBr_02640|M.leprae_Br4923 AGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRTNFFDTYFNNAV
: . . .: . *: ::* * :
Rv3787c|M.tuberculosis_H37Rv AAG---VRQAVILASGLDARGYRLPWPADTTVFEVDQPRVLEFKAQTLAG
Mb3816c|M.bovis_AF2122/97 AAG---VRQAVILASGLDARGYRLPWPADTTVFEVDQPRVLEFKAQTLAG
MSMEG_1888|M.smegmatis_MC2_155 AAG---VRQAVILASGLDARGYRLDWPAGTVLFEIDMPDVLEFKGRALTD
MMAR_3534|M.marinum_M RAG---VRQAVILASGLDARAYRLDWPAGMTVFEIDQPEVIAFKTTTLAG
MUL_2766|M.ulcerans_Agy99 RAG---VRQAVILASGLDARAYRLDWPAGMTVFEIDQPEVIAFKTTTLAG
MAV_2110|M.avium_104 QAG---IRQAVILASGLDARAYRLAWPAGTTVFEIDQPQVIEFKTATLAK
Mflv_5023|M.gilvum_PYR-GCK AAG---VRQAVILASGLDARGYRLTWPEGMTLFEIDQPEVIEFKRTTLAG
Mvan_1346|M.vanbaalenii_PYR-1 RAGPPPIRQGVILASGLDARAYRLSWPQDMVLFEIDQPEVLTFKAATLAD
TH_0301|M.thermoresistible__bu ARG---IRQAVILAAGLDARAYRLRWPDGTVVYEIDQPRVIEFKTRTLAR
MAB_3787|M.abscessus_ATCC_1997 AAG---VRQAVILASGLDTRAYRLTWPAGTTVYELDQPEVITFKTDTLAE
MLBr_02640|M.leprae_Br4923 IDG---IRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLAYKSTTLAE
* :** ****:***:*.*** ** . .::*:* * *: :* :*:
Rv3787c|M.tuberculosis_H37Rv LGAQPTADLRMVPADLRHDWPDALRRGGFDAAEPAAWIAEGLFGYLPPDA
Mb3816c|M.bovis_AF2122/97 LGAQPTADLRMVPADLRHDWPDALRRGGFDAAEPAAWIAEGLFGYLPPDA
MSMEG_1888|M.smegmatis_MC2_155 LRAEPTAEVRMVAVDLRDDWPAALRAKGFDPTRPTAWSAEGLLPFLPPEA
MMAR_3534|M.marinum_M LGAVPRADLRTVAVDLRQDWPKALTDAGFDAGSPTAWIAEGLFGYLPPEA
MUL_2766|M.ulcerans_Agy99 LGAVPRADLRTVAVDLRQDWPKALTEAGFDAGRPTAWIAEGLFGYLPPEA
MAV_2110|M.avium_104 LGATPQATLRTVAVDLRDDWPKALVEAGFDKGQPTAWIAEGLFGYLPPEA
Mflv_5023|M.gilvum_PYR-GCK LGATPSTHLATVAIDLREDWPTALRDAGFDPSVPTAWIAEGLLGYLPPDA
Mvan_1346|M.vanbaalenii_PYR-1 LGVRPSTKLATVAVDLRNDWPAALAEAGFDASAPTAWIAEGLLGYLPPDA
TH_0301|M.thermoresistible__bu LGAEPTAERRTVAADLRHDWPAALTAAGFDPAAPTVWIAEGLFGYLPPDA
MAB_3787|M.abscessus_ATCC_1997 LGASPSAERRTVAIDLREDWPAALQAAGFDPEQPTVWSAEGLLIYLPPEA
MLBr_02640|M.leprae_Br4923 HGVTPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGLLMYLPATA
. * : *. ***.*** ** *** :.* ****: :**. *
Rv3787c|M.tuberculosis_H37Rv QNRLLDHVTDLSAPGSRLALE-AFLGSADRDSARVEEMIRTAT---RGWR
Mb3816c|M.bovis_AF2122/97 QNRLLDHVTDLSAPGSRLALE-AFLGSADRDSARVEEMIRTAT---RGWR
MSMEG_1888|M.smegmatis_MC2_155 QDRLLDAITSLSASGSRLAAEVALLGSDSEDGALGADGARDLEPLLARWR
MMAR_3534|M.marinum_M QDRLLDNITALSAAGSRLACE-AIPNRPQQDAEKARELMRKAT---ARWR
MUL_2766|M.ulcerans_Agy99 QDRLLDNITALSATGSRLACE-AIPNRPQQDAEKARELMRKAT---ARWR
MAV_2110|M.avium_104 QDRLLDNITALSADGSRLACE-AIPDMSEVDTEKAQEMMRRAT---AKWR
Mflv_5023|M.gilvum_PYR-GCK QDRLLDTIGELSASGSRLAVE-SVPSQQSADQDEMREKMKDST---DRWR
Mvan_1346|M.vanbaalenii_PYR-1 QDRLLDTIGELSAPGSRLAVE-SVPTHHEADQEELREKMKESS---DRWR
TH_0301|M.thermoresistible__bu QDRLLDQITDLSAVGSRLGVE-GVPAHADADEDEVRQRMRILA---ERWR
MAB_3787|M.abscessus_ATCC_1997 QDRLFASITELSAPGSRLVCE-QVPGLETADFSKARELTRQFA-------
MLBr_02640|M.leprae_Br4923 QDGLFTEIGGLSAVGSRIAVE-TSPLHGDEWREQMQLRFRRVS----DAL
*: *: : *** ***: * :
Rv3787c|M.tuberculosis_H37Rv EHGFHLDIWALNYAGP-RHEVSGYLDNHGWRSVGTTTAQLLAAHD--LPA
Mb3816c|M.bovis_AF2122/97 EHGFHLDIWALNYAGP-RHEVSGYLDNHGWRSVGTTTAQLLAAHD--LPA
MSMEG_1888|M.smegmatis_MC2_155 EHGFDLDLGDLGNSGP-RNDVDDYLEARGWTSTRTPLAALLDAAG--LEV
MMAR_3534|M.marinum_M EHGFELEFGDLGYEGD-RADVALYLQDLGWQSVGTQMGQLLADNG--AAP
MUL_2766|M.ulcerans_Agy99 EHGFELEFGDLGYEGD-RADVELYLQNLGWQSVGTQMSQLLADNG--AAP
MAV_2110|M.avium_104 EHGFDLEFGDLGYQGE-RNDVAVYLDGLGWRSVGVPMSQLLADAG--LEA
Mflv_5023|M.gilvum_PYR-GCK SHGFDLDFSELVFLGE-RANVPDYLRERGWAVTATPTNDLLTRYG--LAP
Mvan_1346|M.vanbaalenii_PYR-1 SHGFDVDFSELVFLGE-RADVTDYLRDRGWTVDATPTNQLLTRYG--LAP
TH_0301|M.thermoresistible__bu AHGLDLDFSELIYLGD-RADVAGYLADRGWRCTALTANELLVRTG--LPP
MAB_3787|M.abscessus_ATCC_1997 GETLDLDLESLVYTQE-RQMAADWLAEHGWTVITEENDELYARLN--LDP
MLBr_02640|M.leprae_Br4923 GFEQAVDVQELIYHDENRAVVADWLNRHGWRATAQSAPDEMRRVGRWGDG
::. * * . :* ** .
Rv3787c|M.tuberculosis_H37Rv APALPAGLADRP-NYWTCVLG------
Mb3816c|M.bovis_AF2122/97 APALPAGLADRP-NYWTCVLG------
MSMEG_1888|M.smegmatis_MC2_155 PRPADGRKSLSD-NYYSTAIKG-----
MMAR_3534|M.marinum_M IPHNDDSVTMADTIYYSSVLTA-----
MUL_2766|M.ulcerans_Agy99 IPHNNDSVTMADTIYYSSVLTA-----
MAV_2110|M.avium_104 IPQTNDSVSVADTIYYSSVLAK-----
Mflv_5023|M.gilvum_PYR-GCK LDTDEG---FAEVVYVNAAR-------
Mvan_1346|M.vanbaalenii_PYR-1 LDSDEG---FAEVVYVSATR-------
TH_0301|M.thermoresistible__bu AENDGR---VTEVVYLSSEKSPGKRAG
MAB_3787|M.abscessus_ATCC_1997 PNPLLRS-IFPNIVYVDATLG------
MLBr_02640|M.leprae_Br4923 VPMADDKDAFAEFVTAHRL--------