For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRTDNDSWDLATSVGATATMVAAARAIATNADNPLIADRFAEPLVRAVGVDFFTRWVTGDLVVADVDDTE SGWQLAQMPDAMAVRTRFFDAFFQDATRAGVRQAVILASGLDARAYRLDWPAGMTVFEIDQPEVIAFKTT TLAGLGAVPRADLRTVAVDLRQDWPKALTDAGFDAGSPTAWIAEGLFGYLPPEAQDRLLDNITALSAAGS RLACEAIPNRPQQDAEKARELMRKATARWREHGFELEFGDLGYEGDRADVALYLQDLGWQSVGTQMGQLL ADNGAAPIPHNDDSVTMADTIYYSSVLTA*
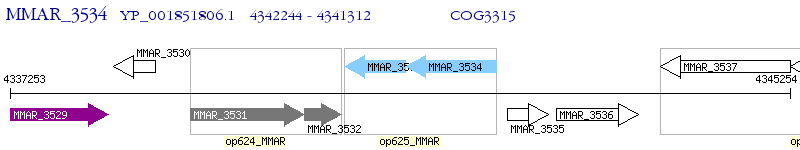
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3534 | - | - | 100% (310) | O-methyltransferase |
| M. marinum M | MMAR_1059 | - | 4e-96 | 59.42% (308) | O-methyltransferase |
| M. marinum M | MMAR_1069 | - | 6e-90 | 55.84% (308) | O-methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3816c | - | 1e-100 | 63.89% (288) | hypothetical protein Mb3816c |
| M. gilvum PYR-GCK | Mflv_5023 | - | 1e-102 | 60.06% (308) | putative methyltransferase |
| M. tuberculosis H37Rv | Rv3787c | - | 1e-100 | 63.89% (288) | hypothetical protein Rv3787c |
| M. leprae Br4923 | MLBr_02640 | - | 8e-61 | 45.54% (303) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_3787 | - | 3e-78 | 54.77% (283) | hypothetical protein MAB_3787 |
| M. avium 104 | MAV_2110 | - | 1e-144 | 80.52% (308) | methyltransferase, putative, family protein |
| M. smegmatis MC2 155 | MSMEG_1482 | - | 1e-98 | 60.46% (306) | methyltransferase |
| M. thermoresistible (build 8) | TH_2706 | - | 2e-85 | 59.79% (286) | PUTATIVE putative methyltransferase |
| M. ulcerans Agy99 | MUL_2766 | - | 1e-175 | 97.42% (310) | O-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1346 | - | 1e-100 | 59.81% (311) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3534|M.marinum_M MPRTDNDSWDLATSVGATATMVAAARAIATNADNPLIADRFAEPLVRAVG
MUL_2766|M.ulcerans_Agy99 MPRTDNDSWDLATSVGATATMVAAARAIATNADNPLIADRFAEPLVRAVG
MAV_2110|M.avium_104 MPRTDNDTWDLSTSVGATATMVAAARAIATNADNPLIEDRFAEPLVRAVG
Mb3816c|M.bovis_AF2122/97 MARTDDDSWDLATGVGATATLVAAGRARAARAAQPLIDDPFAEPLVRAVG
Rv3787c|M.tuberculosis_H37Rv MARTDDDSWDLATGVGATATLVAAGRARAARAAQPLIDDPFAEPLVRAVG
Mflv_5023|M.gilvum_PYR-GCK MPRTENDTWDLASSVGATATMVAAARAVATRAEDPVIDDPYAEPLVRAVG
Mvan_1346|M.vanbaalenii_PYR-1 MARTDNDTWDLASSVGATATMVAAARAVATRAPRPVIDDPFAAPLVQAVG
MSMEG_1482|M.smegmatis_MC2_155 MARTENDTWDLASSVGATATMVAAARALATNADEPAIDDPFAAPLVRAVG
TH_2706|M.thermoresistible__bu MARSESDTWDLTSGVGVTATMVAAARALASREPEPLIDDPFAAPLVRAVG
MAB_3787|M.abscessus_ATCC_1997 MARTDDDSWDLASSVGATATMVAAQRALASHVPNPLINDPYAEPLVRAVG
MLBr_02640|M.leprae_Br4923 -MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLLVTNTG
*:..*:**: :.**.**.:*** ** : . * * :* ** .*
MMAR_3534|M.marinum_M VDFFTRWVTGDLVVADVDDT--ESGWQLAQMPDAMAVRTRFFDAFFQDAT
MUL_2766|M.ulcerans_Agy99 VDFFTRWVTGDLVVADVDDT--ESGWQLAQMPDAMAVRARFFDAFFQDAT
MAV_2110|M.avium_104 VDFFTRWVTGDLVAADVDDH--DSGWKLEHMPVAMAARTRFFDSFFQGAT
Mb3816c|M.bovis_AF2122/97 VEFLTRWATGELDAADVDDP--DAAWGLQRMTTELVVRTRYFDQFFLDAA
Rv3787c|M.tuberculosis_H37Rv VEFLTRWATGELDAADVDDP--DAAWGLQRMTTELVVRTRYFDQFFLDAA
Mflv_5023|M.gilvum_PYR-GCK IDFFAKLASGELTPEDLDVSGDESPVGMSRFADGMAARTRFFDDFFTDAC
Mvan_1346|M.vanbaalenii_PYR-1 VDFFTRLATGELTTEELDTNDAESPVGMSRFADAMAARTRFFDDFFAEAM
MSMEG_1482|M.smegmatis_MC2_155 VDFFTKLVDDNFDLTALDP---EAAEGLTRFANGMAARTRFFDDFFLDAA
TH_2706|M.thermoresistible__bu VEFFTRLVDGDLELP-ADTA--EERAAAQVLTEVMAVRTRFFDDFFTDAT
MAB_3787|M.abscessus_ATCC_1997 IEYFTKLASGAFDPEQMAGL-VQLGITADGMANGMASRTWYYDTAFESST
MLBr_02640|M.leprae_Br4923 AGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRTNFFDTYFNNAV
: . . : : . *: ::* * :
MMAR_3534|M.marinum_M RAG---VRQAVILASGLDARAYRLDWPAGMTVFEIDQPEVIAFKTTTLAG
MUL_2766|M.ulcerans_Agy99 RAG---VRQAVILASGLDARAYRLDWPAGMTVFEIDQPEVIAFKTTTLAG
MAV_2110|M.avium_104 QAG---IRQAVILASGLDARAYRLAWPAGTTVFEIDQPQVIEFKTATLAK
Mb3816c|M.bovis_AF2122/97 AAG---VRQAVILASGLDARGYRLPWPADTTVFEVDQPRVLEFKAQTLAG
Rv3787c|M.tuberculosis_H37Rv AAG---VRQAVILASGLDARGYRLPWPADTTVFEVDQPRVLEFKAQTLAG
Mflv_5023|M.gilvum_PYR-GCK AAG---VRQAVILASGLDARGYRLTWPEGMTLFEIDQPEVIEFKRTTLAG
Mvan_1346|M.vanbaalenii_PYR-1 RAGPPPIRQGVILASGLDARAYRLSWPQDMVLFEIDQPEVLTFKAATLAD
MSMEG_1482|M.smegmatis_MC2_155 RAG---VRQFVILASGLDARAYRLPWPAGSVVFEIDQPDVIEFKTKALAD
TH_2706|M.thermoresistible__bu RVG---ARQAVILASGLDARAYRLTWPDGTVVYEIDQPEVIAFKTRTLAE
MAB_3787|M.abscessus_ATCC_1997 AAG---VRQAVILASGLDTRAYRLTWPAGTTVYELDQPEVITFKTDTLAE
MLBr_02640|M.leprae_Br4923 IDG---IRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLAYKSTTLAE
* ** ********:*.*** ** . .::*:*** *: :* :**
MMAR_3534|M.marinum_M LGAVPRADLRTVAVDLRQDWPKALTDAGFDAGSPTAWIAEGLFGYLPPEA
MUL_2766|M.ulcerans_Agy99 LGAVPRADLRTVAVDLRQDWPKALTEAGFDAGRPTAWIAEGLFGYLPPEA
MAV_2110|M.avium_104 LGATPQATLRTVAVDLRDDWPKALVEAGFDKGQPTAWIAEGLFGYLPPEA
Mb3816c|M.bovis_AF2122/97 LGAQPTADLRMVPADLRHDWPDALRRGGFDAAEPAAWIAEGLFGYLPPDA
Rv3787c|M.tuberculosis_H37Rv LGAQPTADLRMVPADLRHDWPDALRRGGFDAAEPAAWIAEGLFGYLPPDA
Mflv_5023|M.gilvum_PYR-GCK LGATPSTHLATVAIDLREDWPTALRDAGFDPSVPTAWIAEGLLGYLPPDA
Mvan_1346|M.vanbaalenii_PYR-1 LGVRPSTKLATVAVDLRNDWPAALAEAGFDASAPTAWIAEGLLGYLPPDA
MSMEG_1482|M.smegmatis_MC2_155 LGALPTTDRRAVAVDLRFDWPAALTEAGFDPAEPTAWIAEGLLGYLPPEA
TH_2706|M.thermoresistible__bu LGAPPTAPHRPVAVDLRDDWPAALTAAGFDPTVATAWSAEGLLRYLPPDA
MAB_3787|M.abscessus_ATCC_1997 LGASPSAERRTVAIDLREDWPAALQAAGFDPEQPTVWSAEGLLIYLPPEA
MLBr_02640|M.leprae_Br4923 HGVTPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGLLMYLPATA
*. * : *. *** *** ** .*** :.* ****: ***. *
MMAR_3534|M.marinum_M QDRLLDNITALSAAGSRLACEAIPNRPQQDAEKARELMRKATARWREHGF
MUL_2766|M.ulcerans_Agy99 QDRLLDNITALSATGSRLACEAIPNRPQQDAEKARELMRKATARWREHGF
MAV_2110|M.avium_104 QDRLLDNITALSADGSRLACEAIPDMSEVDTEKAQEMMRRATAKWREHGF
Mb3816c|M.bovis_AF2122/97 QNRLLDHVTDLSAPGSRLALEAFLGSADRDSARVEEMIRTATRGWREHGF
Rv3787c|M.tuberculosis_H37Rv QNRLLDHVTDLSAPGSRLALEAFLGSADRDSARVEEMIRTATRGWREHGF
Mflv_5023|M.gilvum_PYR-GCK QDRLLDTIGELSASGSRLAVESVPSQQSADQDEMREKMKDSTDRWRSHGF
Mvan_1346|M.vanbaalenii_PYR-1 QDRLLDTIGELSAPGSRLAVESVPTHHEADQEELREKMKESSDRWRSHGF
MSMEG_1482|M.smegmatis_MC2_155 QDRLLDQIAELSAPGSRVAVEEIPAIEEEDHEEIAARMKDFSDRWRAHGF
TH_2706|M.thermoresistible__bu QDRLFDHITDLSAPGSRLATEDHPGPGAGLGEKADELGR----RWREHGF
MAB_3787|M.abscessus_ATCC_1997 QDRLFASITELSAPGSRLVCEQVPGLETADFSKARELTR----QFAGETL
MLBr_02640|M.leprae_Br4923 QDGLFTEIGGLSAVGSRIAVETSPLHGDEWREQMQLRFRRVS-DALGFEQ
*: *: : *** ***:. * . :
MMAR_3534|M.marinum_M ELEFGDLGYEGD-RADVALYLQDLGWQSVGTQMGQLLADNG-----AAPI
MUL_2766|M.ulcerans_Agy99 ELEFGDLGYEGD-RADVELYLQNLGWQSVGTQMSQLLADNG-----AAPI
MAV_2110|M.avium_104 DLEFGDLGYQGE-RNDVAVYLDGLGWRSVGVPMSQLLADAG-----LEAI
Mb3816c|M.bovis_AF2122/97 HLDIWALNYAGP-RHEVSGYLDNHGWRSVGTTTAQLLAAHD-----LPAA
Rv3787c|M.tuberculosis_H37Rv HLDIWALNYAGP-RHEVSGYLDNHGWRSVGTTTAQLLAAHD-----LPAA
Mflv_5023|M.gilvum_PYR-GCK DLDFSELVFLGE-RANVPDYLRERGWAVTATPTNDLLTRYG-----LAPL
Mvan_1346|M.vanbaalenii_PYR-1 DVDFSELVFLGE-RADVTDYLRDRGWTVDATPTNQLLTRYG-----LAPL
MSMEG_1482|M.smegmatis_MC2_155 DLDFTELVFLGD-RADVSSYLQGHGWKTTSMTSDELLVHNG-----LPRV
TH_2706|M.thermoresistible__bu EVQLSELFHPGE-RTPVVDYLRGRGWDARARPRAEVFAGYGREFPDSAAL
MAB_3787|M.abscessus_ATCC_1997 DLDLESLVYTQE-RQMAADWLAEHGWTVITEENDELYARLN-----LDPP
MLBr_02640|M.leprae_Br4923 AVDVQELIYHDENRAVVADWLNRHGWRATAQSAPDEMRRVG---RWGDGV
::. * . * . :* ** : .
MMAR_3534|M.marinum_M PHNDDSVTMADT-IYYSSVLTA
MUL_2766|M.ulcerans_Agy99 PHNNDSVTMADT-IYYSSVLTA
MAV_2110|M.avium_104 PQTNDSVSVADT-IYYSSVLAK
Mb3816c|M.bovis_AF2122/97 PALP--AGLADRPNYWTCVLG-
Rv3787c|M.tuberculosis_H37Rv PALP--AGLADRPNYWTCVLG-
Mflv_5023|M.gilvum_PYR-GCK DTDE---GFAEV-VYVNAAR--
Mvan_1346|M.vanbaalenii_PYR-1 DSDE---GFAEV-VYVSATR--
MSMEG_1482|M.smegmatis_MC2_155 DDDA---QIGSV-VYVTATR--
TH_2706|M.thermoresistible__bu AAMRDSSAVTAVRPVVRAGMR-
MAB_3787|M.abscessus_ATCC_1997 NPLLR--SIFPNIVYVDATLG-
MLBr_02640|M.leprae_Br4923 PMAD---DKDAFAEFVTAHRL-
.