For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MMISTDQTRSGKFTVTPRRDGDLLVLHVTGDLDVLTAPTLSTHLDIALADAPPVVIVDISDVSFLSSAGI GLLVETHRLTERGGISLRVVADGAATSRPLRMMRIDEVIDLYATTDEAKAGRPA*
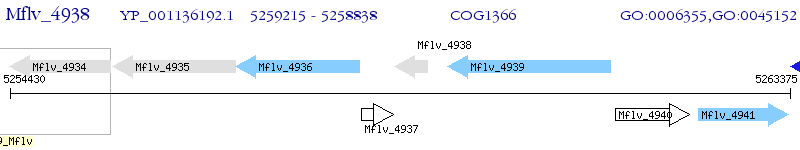
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4938 | - | - | 100% (125) | anti-sigma-factor antagonist |
| M. gilvum PYR-GCK | Mflv_3284 | - | 1e-19 | 43.48% (115) | anti-sigma-factor antagonist |
| M. gilvum PYR-GCK | Mflv_2021 | - | 1e-14 | 39.05% (105) | anti-sigma-factor antagonist |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3712c | - | 2e-08 | 36.84% (76) | hypothetical protein Mb3712c |
| M. tuberculosis H37Rv | Rv3687c | rsfB | 4e-13 | 34.55% (110) | anti-anti-sigma factor RSFB (anti-sigma factor antagonist) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0214 | - | 1e-18 | 42.86% (119) | hypothetical protein MMAR_0214 |
| M. avium 104 | MAV_5272 | - | 2e-16 | 41.51% (106) | anti-anti-sigma factor |
| M. smegmatis MC2 155 | MSMEG_6127 | - | 2e-21 | 50.48% (105) | anti-anti-sigma factor |
| M. thermoresistible (build 8) | TH_0498 | - | 2e-18 | 42.45% (106) | anti-anti-sigma factor |
| M. ulcerans Agy99 | MUL_3602 | - | 1e-10 | 32.71% (107) | anti sigma factor antagonist |
| M. vanbaalenii PYR-1 | Mvan_1481 | - | 4e-41 | 71.43% (119) | anti-sigma-factor antagonist |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3712c|M.bovis_AF2122/97 ------------------------------------------------MA
Rv3687c|M.tuberculosis_H37Rv -------MSAPDSITVTVADHNGVAVLSIGGEIDLITAAALEEAIGEVVA
Mflv_4938|M.gilvum_PYR-GCK MMMISTDQTRSGKFTVTPRRDGDLLVLHVTGDLDVLTAPTLSTHLDIALA
Mvan_1481|M.vanbaalenii_PYR-1 --MDVSGHGRSRGFSVSPDRQGAALVLRVTGDLDAMTAPTLVTHLDIALA
MSMEG_6127|M.smegmatis_MC2_155 MTSQ-----DPANCTVEERRVGDITVVAVTGTVDMLTAPKLEDAIGSAAK
TH_0498|M.thermoresistible__bu --------------VIDERWDADIAVVSATGVVDMLSAPQLDEAIRTALG
MMAR_0214|M.marinum_M MVEQSGDPTPPTAFGIEKSRVGEAVVVTVSGELDMLTAPRLAEMLRTSLI
MAV_5272|M.avium_104 MGEQPGDLADPTPFEVGKHQVDQAVVLTVSGEVDMLSSPMLAEAVQTALA
MUL_3602|M.ulcerans_Agy99 ----------MDLLAVEHEVRQDTVVVRAKGDVDSSTVEQLVVHLAAAMQ
Mb3712c|M.bovis_AF2122/97 DNP----TALVIDLSAVEFLGSVGLKILAATSEKIGQS-VKFGVVARGSV
Rv3687c|M.tuberculosis_H37Rv DNP----TALVIDLSAVEFLGSVGLKILAATSEKIGQS-VKFGVVARGSV
Mflv_4938|M.gilvum_PYR-GCK DAP----PVVIVDISDVSFLSSAGIGLLVETHRLTERGGISLRVVADGAA
Mvan_1481|M.vanbaalenii_PYR-1 DAP----PVVVVDMTDVEFLSSAGISVLVETHRLAERADISLRVVADGPA
MSMEG_6127|M.smegmatis_MC2_155 SEP----SAVVVDLSAVDFLASAGMGVLVAAHGELAPA-VRLVVVADGPA
TH_0498|M.thermoresistible__bu KKP----RALVVDFTAVDFLASAGMGVLVAIHDEVSPA-VNLTIVADGPA
MMAR_0214|M.marinum_M EAP----ATLIVDLSKVDFLASAGMTVLVSAKAQAVAP-TRFVVVADGPA
MAV_5272|M.avium_104 AKP----AALIVDLSKVGFLASAGMTVLVTTQAELQPP-TKFAVVADGSA
MUL_3602|M.ulcerans_Agy99 LAATHPARLVVIDLQPVTFFGSAGLNAVLECHEQGMAAGTSVRLVADHGQ
. :::*: * *:.*.*: : . :**
Mb3712c|M.bovis_AF2122/97 TRRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------
Rv3687c|M.tuberculosis_H37Rv TRRPIHLMGLDKTFRLFSTLHDALTGVRGGRIDR-------
Mflv_4938|M.gilvum_PYR-GCK TSRPLRMMRIDEVIDLYATTDEAK----AGRPA--------
Mvan_1481|M.vanbaalenii_PYR-1 TSRPFRMMRLDEVIDLYPTLADAMGERQQGRPPT-------
MSMEG_6127|M.smegmatis_MC2_155 TSRPLKLVGIADVVDLFATLDEALSSLKT------------
TH_0498|M.thermoresistible__bu TSRPLKLLGIADLVPLHATLDEALAAVAT------------
MMAR_0214|M.marinum_M TSRPIRLMGIDAMLPLYDALDDALNDISDS-----------
MAV_5272|M.avium_104 TSRPIKLMGIDSVLSLYPSLNDALSALSGA-----------
MUL_3602|M.ulcerans_Agy99 LLAPIQVTELDRIFEIYPTLAGALRPGKPGEHDLEDVERPR
*::: : . :. : *