For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRTNNDAWDLATSVGATATMVAAARAVATRADNPLIDDPFAEPLVRAVGIDFFTRWAAGNIKATDVDDPD GTWGLQRLADLLAARTRYFDAFFRDATSAGIRQAVILASGLDARAYR*
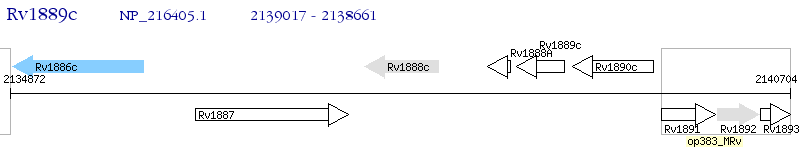
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1889c | - | - | 100% (118) | hypothetical protein Rv1889c |
| M. tuberculosis H37Rv | Rv3787c | - | 6e-46 | 72.41% (116) | hypothetical protein Rv3787c |
| M. tuberculosis H37Rv | Rv3399 | - | 8e-39 | 64.17% (120) | hypothetical protein Rv3399 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1922c | - | 2e-64 | 100.00% (118) | hypothetical protein Mb1922c |
| M. gilvum PYR-GCK | Mflv_5023 | - | 2e-42 | 69.17% (120) | putative methyltransferase |
| M. leprae Br4923 | MLBr_02640 | - | 6e-21 | 44.17% (120) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_3787 | - | 4e-33 | 57.60% (125) | hypothetical protein MAB_3787 |
| M. marinum M | MMAR_3534 | - | 3e-48 | 74.58% (118) | O-methyltransferase |
| M. avium 104 | MAV_2110 | - | 6e-48 | 74.58% (118) | methyltransferase, putative, family protein |
| M. smegmatis MC2 155 | MSMEG_1888 | - | 7e-43 | 73.04% (115) | methyltransferase |
| M. thermoresistible (build 8) | TH_2706 | - | 2e-34 | 60.17% (118) | PUTATIVE putative methyltransferase |
| M. ulcerans Agy99 | MUL_2766 | - | 7e-48 | 73.73% (118) | O-methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1346 | - | 2e-39 | 66.67% (123) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1889c|M.tuberculosis_H37Rv MPRTNNDAWDLATSVGATATMVAAARAVATRADNPLIDDPFAEPLVRAVG
Mb1922c|M.bovis_AF2122/97 MPRTNNDAWDLATSVGATATMVAAARAVATRADNPLIDDPFAEPLVRAVG
MSMEG_1888|M.smegmatis_MC2_155 MEST-HETWDLATSVGATATMVAAGRARATTTG--LIDDRFAEPLVRAVG
MMAR_3534|M.marinum_M MPRTDNDSWDLATSVGATATMVAAARAIATNADNPLIADRFAEPLVRAVG
MUL_2766|M.ulcerans_Agy99 MPRTDNDSWDLATSVGATATMVAAARAIATNADNPLIADRFAEPLVRAVG
MAV_2110|M.avium_104 MPRTDNDTWDLSTSVGATATMVAAARAIATNADNPLIEDRFAEPLVRAVG
Mflv_5023|M.gilvum_PYR-GCK MPRTENDTWDLASSVGATATMVAAARAVATRAEDPVIDDPYAEPLVRAVG
Mvan_1346|M.vanbaalenii_PYR-1 MARTDNDTWDLASSVGATATMVAAARAVATRAPRPVIDDPFAAPLVQAVG
TH_2706|M.thermoresistible__bu MARSESDTWDLTSGVGVTATMVAAARALASREPEPLIDDPFAAPLVRAVG
MAB_3787|M.abscessus_ATCC_1997 MARTDDDSWDLASSVGATATMVAAQRALASHVPNPLINDPYAEPLVRAVG
MLBr_02640|M.leprae_Br4923 -MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLLVTNTG
: ::**: :.**.**.**** ** : :* * :* ** .*
Rv1889c|M.tuberculosis_H37Rv IDFFTRWAAGNIKATDVDDP--DGTWGLQRLADLLAARTRYFDAFFRDAT
Mb1922c|M.bovis_AF2122/97 IDFFTRWAAGNIKATDVDDP--DGTWGLQRLADLLAARTRYFDAFFRDAT
MSMEG_1888|M.smegmatis_MC2_155 IDFMTRWATGDLAAADVDIP--GATWGMQQMTDLLAARTRYFDAFFGDAA
MMAR_3534|M.marinum_M VDFFTRWVTGDLVVADVDDT--ESGWQLAQMPDAMAVRTRFFDAFFQDAT
MUL_2766|M.ulcerans_Agy99 VDFFTRWVTGDLVVADVDDT--ESGWQLAQMPDAMAVRARFFDAFFQDAT
MAV_2110|M.avium_104 VDFFTRWVTGDLVAADVDDH--DSGWKLEHMPVAMAARTRFFDSFFQGAT
Mflv_5023|M.gilvum_PYR-GCK IDFFAKLASGELTPEDLDVSGDESPVGMSRFADGMAARTRFFDDFFTDAC
Mvan_1346|M.vanbaalenii_PYR-1 VDFFTRLATGELTTEELDTNDAESPVGMSRFADAMAARTRFFDDFFAEAM
TH_2706|M.thermoresistible__bu VEFFTRLVDGDLELP-ADTA--EERAAAQVLTEVMAVRTRFFDDFFTDAT
MAB_3787|M.abscessus_ATCC_1997 IEYFTKLASGAFDPEQMAGL-VQLGITADGMANGMASRTWYYDTAFESST
MLBr_02640|M.leprae_Br4923 AGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRTNFFDTYFNNAV
: . . : * *: ::* * :
Rv1889c|M.tuberculosis_H37Rv SAG---IRQAVILASGLDARAYR---------------------------
Mb1922c|M.bovis_AF2122/97 SAG---IRQAVILASGLDARAYR---------------------------
MSMEG_1888|M.smegmatis_MC2_155 AAG---VRQAVILASGLDARGYRLDWPAGTVLFEIDMPDVLEFKGRALTD
MMAR_3534|M.marinum_M RAG---VRQAVILASGLDARAYRLDWPAGMTVFEIDQPEVIAFKTTTLAG
MUL_2766|M.ulcerans_Agy99 RAG---VRQAVILASGLDARAYRLDWPAGMTVFEIDQPEVIAFKTTTLAG
MAV_2110|M.avium_104 QAG---IRQAVILASGLDARAYRLAWPAGTTVFEIDQPQVIEFKTATLAK
Mflv_5023|M.gilvum_PYR-GCK AAG---VRQAVILASGLDARGYRLTWPEGMTLFEIDQPEVIEFKRTTLAG
Mvan_1346|M.vanbaalenii_PYR-1 RAGPPPIRQGVILASGLDARAYRLSWPQDMVLFEIDQPEVLTFKAATLAD
TH_2706|M.thermoresistible__bu RVG---ARQAVILASGLDARAYRLTWPDGTVVYEIDQPEVIAFKTRTLAE
MAB_3787|M.abscessus_ATCC_1997 AAG---VRQAVILASGLDTRAYRLTWPAGTTVYELDQPEVITFKTDTLAE
MLBr_02640|M.leprae_Br4923 IDG---IRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLAYKSTTLAE
* ** ********:*.**
Rv1889c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1922c|M.bovis_AF2122/97 --------------------------------------------------
MSMEG_1888|M.smegmatis_MC2_155 LRAEPTAEVRMVAVDLRDDWPAALRAKGFDPTRPTAWSAEGLLPFLPPEA
MMAR_3534|M.marinum_M LGAVPRADLRTVAVDLRQDWPKALTDAGFDAGSPTAWIAEGLFGYLPPEA
MUL_2766|M.ulcerans_Agy99 LGAVPRADLRTVAVDLRQDWPKALTEAGFDAGRPTAWIAEGLFGYLPPEA
MAV_2110|M.avium_104 LGATPQATLRTVAVDLRDDWPKALVEAGFDKGQPTAWIAEGLFGYLPPEA
Mflv_5023|M.gilvum_PYR-GCK LGATPSTHLATVAIDLREDWPTALRDAGFDPSVPTAWIAEGLLGYLPPDA
Mvan_1346|M.vanbaalenii_PYR-1 LGVRPSTKLATVAVDLRNDWPAALAEAGFDASAPTAWIAEGLLGYLPPDA
TH_2706|M.thermoresistible__bu LGAPPTAPHRPVAVDLRDDWPAALTAAGFDPTVATAWSAEGLLRYLPPDA
MAB_3787|M.abscessus_ATCC_1997 LGASPSAERRTVAIDLREDWPAALQAAGFDPEQPTVWSAEGLLIYLPPEA
MLBr_02640|M.leprae_Br4923 HGVTPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGLLMYLPATA
Rv1889c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1922c|M.bovis_AF2122/97 --------------------------------------------------
MSMEG_1888|M.smegmatis_MC2_155 QDRLLDAITSLSASGSRLAAEVALLGSDSEDGALGADGARDLEP-LLARW
MMAR_3534|M.marinum_M QDRLLDNITALSAAGSRLACEAIPNRPQQD-----AEKARELMRKATARW
MUL_2766|M.ulcerans_Agy99 QDRLLDNITALSATGSRLACEAIPNRPQQD-----AEKARELMRKATARW
MAV_2110|M.avium_104 QDRLLDNITALSADGSRLACEAIPDMSEVD-----TEKAQEMMRRATAKW
Mflv_5023|M.gilvum_PYR-GCK QDRLLDTIGELSASGSRLAVESVPSQQSAD-----QDEMREKMKDSTDRW
Mvan_1346|M.vanbaalenii_PYR-1 QDRLLDTIGELSAPGSRLAVESVPTHHEAD-----QEELREKMKESSDRW
TH_2706|M.thermoresistible__bu QDRLFDHITDLSAPGSRLATEDHPGP---------GAGLGEKADELGRRW
MAB_3787|M.abscessus_ATCC_1997 QDRLFASITELSAPGSRLVCEQVPGLETAD---------FSKARELTRQF
MLBr_02640|M.leprae_Br4923 QDGLFTEIGGLSAVGSRIAVETSPLHGDEWR-----EQMQLRFRRVSDAL
Rv1889c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1922c|M.bovis_AF2122/97 --------------------------------------------------
MSMEG_1888|M.smegmatis_MC2_155 REHGFDLDLGDLGNSGP-RNDVDDYLEARGWTSTRTPLAALLDAAGLE-V
MMAR_3534|M.marinum_M REHGFELEFGDLGYEGD-RADVALYLQDLGWQSVGTQMGQLLADNGAAPI
MUL_2766|M.ulcerans_Agy99 REHGFELEFGDLGYEGD-RADVELYLQNLGWQSVGTQMSQLLADNGAAPI
MAV_2110|M.avium_104 REHGFDLEFGDLGYQGE-RNDVAVYLDGLGWRSVGVPMSQLLADAGLEAI
Mflv_5023|M.gilvum_PYR-GCK RSHGFDLDFSELVFLGE-RANVPDYLRERGWAVTATPTNDLLTRYGLAPL
Mvan_1346|M.vanbaalenii_PYR-1 RSHGFDVDFSELVFLGE-RADVTDYLRDRGWTVDATPTNQLLTRYGLAPL
TH_2706|M.thermoresistible__bu REHGFEVQLSELFHPGE-RTPVVDYLRGRGWDARARPRAEVFAGYGRE-F
MAB_3787|M.abscessus_ATCC_1997 AGETLDLDLESLVYTQE-RQMAADWLAEHGWTVITEENDELYARLNLD-P
MLBr_02640|M.leprae_Br4923 G-FEQAVDVQELIYHDENRAVVADWLNRHGWRATAQSAPDEMRRVGRWGD
Rv1889c|M.tuberculosis_H37Rv ---------------------------
Mb1922c|M.bovis_AF2122/97 ---------------------------
MSMEG_1888|M.smegmatis_MC2_155 PRPADGRKSLSDNYYSTAIKG------
MMAR_3534|M.marinum_M PHNDDSVTMADTIYYSSVLTA------
MUL_2766|M.ulcerans_Agy99 PHNNDSVTMADTIYYSSVLTA------
MAV_2110|M.avium_104 PQTNDSVSVADTIYYSSVLAK------
Mflv_5023|M.gilvum_PYR-GCK DTDEG---FAEVVYVNAAR--------
Mvan_1346|M.vanbaalenii_PYR-1 DSDEG---FAEVVYVSATR--------
TH_2706|M.thermoresistible__bu PDSAALAAMRDSSAVTAVRPVVRAGMR
MAB_3787|M.abscessus_ATCC_1997 PNPLLRSIFPNIVYVDATLG-------
MLBr_02640|M.leprae_Br4923 GVPMADDKDAFAEFVTAHRL-------