For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPRGCAGARFACNACLNFLAGLGISEPISPGWAAMERLSGLDAFFLYMETPSQPLNVCCVLELDTSTMPG GYTYGRFHAALEKYVKAAPEFRMKLADTELNLDHPVWVDDDNFQIRHHLRRVAMPAPGGRRELAEICGYI AGLPLDRDRPLWEMWVIEGGARSDTVAVMLKVHHAVVDGVAGANLLSHLCSLQPDAPAPQPVRGTGGGNV LQIAASGLEGFASRPVRLATVVPATVLTLVRTLLRAREGRTMAAPFSAPPTPFNGPLGRLRNIAYTQLDM RDVKRVKDRFGVTINDVVVALCAGALRRFLLEHGVLPEAPLVATVPVSVHDKSDRPGRNQATWMFCRVPS QISDPAQRIRTIAAGNTVAKDHAAAIGPTLLHDWIQFGGSTMFGAAMRILPHISITHSPAYNLILSNVPG PQAQLYFLGCRMDSMFPLGPLLGNAGLNITVMSLNGELGVGIVSCPDLLPDLWGVADGFPEALKELLECS DDQPEGSNHQDS
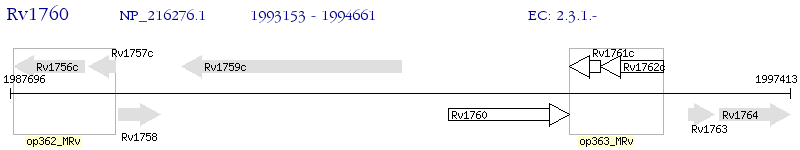
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1760 | - | - | 100% (502) | hypothetical protein Rv1760 |
| M. tuberculosis H37Rv | Rv1425 | - | 6e-70 | 35.87% (460) | hypothetical protein Rv1425 |
| M. tuberculosis H37Rv | Rv3740c | - | 4e-56 | 33.05% (463) | hypothetical protein Rv3740c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1791 | - | 0.0 | 99.55% (442) | hypothetical protein Mb1791 |
| M. gilvum PYR-GCK | Mflv_5212 | - | 1e-138 | 53.49% (458) | hypothetical protein Mflv_5212 |
| M. leprae Br4923 | MLBr_01244 | - | 4e-28 | 25.71% (455) | hypothetical protein MLBr_01244 |
| M. abscessus ATCC 19977 | MAB_1917 | - | 1e-152 | 56.17% (454) | hypothetical protein MAB_1917 |
| M. marinum M | MMAR_2634 | - | 0.0 | 79.65% (457) | hypothetical protein MMAR_2634 |
| M. avium 104 | MAV_3015 | - | 1e-175 | 64.94% (445) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. smegmatis MC2 155 | MSMEG_4297 | - | 1e-160 | 59.74% (462) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. thermoresistible (build 8) | TH_2477 | - | 1e-157 | 57.61% (460) | PUTATIVE acyltransferase, ws/dgat/mgat subfamily protein |
| M. ulcerans Agy99 | MUL_1818 | - | 2e-69 | 35.43% (460) | hypothetical protein MUL_1818 |
| M. vanbaalenii PYR-1 | Mvan_3601 | - | 1e-162 | 59.74% (462) | hypothetical protein Mvan_3601 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1760|M.tuberculosis_H37Rv MPRGCAGARFACNACLNFLAGLGISEPISPGWAAMERLSGLDAFFLYMET
Mb1791|M.bovis_AF2122/97 MPRGCAGARFACNACLNFLAGLGISEPISPGWAAMERLSGLDAFFLYMET
MMAR_2634|M.marinum_M ----------------------------------MERLSGLDAFFLYLET
MAV_3015|M.avium_104 ---------------------------------------------MYLES
MSMEG_4297|M.smegmatis_MC2_155 ----------------------------------MQRLSGLDASFLYLET
Mvan_3601|M.vanbaalenii_PYR-1 -------------------MDLLTPAPPRHTIGGMQRLSGLDASFLYLES
TH_2477|M.thermoresistible__bu -------------------------VPPPYDRG-MQRLSGLDASFLYLET
MAB_1917|M.abscessus_ATCC_1997 ----------------------------------MQRLSGLDASFLYLET
Mflv_5212|M.gilvum_PYR-GCK ----------------------------------MERLSGLDAGMLYSES
MUL_1818|M.ulcerans_Agy99 ----------------------------------MRRLSSVDAAFWSAET
MLBr_01244|M.leprae_Br4923 ------------------------MADSVEGIGPFDELGALDYLLHRGEA
*:
Rv1760|M.tuberculosis_H37Rv -PSQPLNVCCVLELDTSTMPGGYTYGRFHAALEKYVKAAPEFRMKLADTE
Mb1791|M.bovis_AF2122/97 -PSQPLNVCCVLELDTSTMPGGYTYGRFHAALEKYVKAAPEFRMKLADTE
MMAR_2634|M.marinum_M -PAQPMNVCCVLELDTSTMPGGYRYKRFHAALARHLQAVPEFRMKLADTQ
MAV_3015|M.avium_104 -PTQLLNICCVMELDPATMPGGYTFERFRAALSRQLEAVPEFRLKLADTP
MSMEG_4297|M.smegmatis_MC2_155 -AAQPMHVCSVLELDTSTMPGGYTFDRLRDALSLRIKAMPEFREKLADSR
Mvan_3601|M.vanbaalenii_PYR-1 -AAQPLHVCSILELDTSTMPGGYTFDRLRDGLALRIKAMPEFREKIADSR
TH_2477|M.thermoresistible__bu -PDQPLHVCSILELDTSTIPGGYDFHQLREALLLRIKAMPEFREKLADNR
MAB_1917|M.abscessus_ATCC_1997 -PTQPMHVCGVLELDTTTIPGGYSFEKLRTKLAERVEGIPSFREKLADSR
Mflv_5212|M.gilvum_PYR-GCK -PTVPLHVCSVTELDTSTVPGGYTFAKFRDHLASRVPAIPEFRTKLADND
MUL_1818|M.ulcerans_Agy99 -AGWHMHVGALAICDPTDAP-DYSFQRLRELLIERLPEIPQLRWRVTGAP
MLBr_01244|M.leprae_Br4923 NPRTRAGIMAVELLDT-----TPDWNRFRSRIEDVSQRVLRLRQKVVVPT
. : : *. : ::: : :* ::.
Rv1760|M.tuberculosis_H37Rv LNLDHPVWVDDDNFQIRHHLRRVAMPAPGGRRELAEICGYIAGLPLDRDR
Mb1791|M.bovis_AF2122/97 LNLDHPVWVDDDNFQIRHHLRRVAMPAPGGRRELAEICGYIAGLPLDRDR
MMAR_2634|M.marinum_M LNLDHPVWVDDEGFQLRRHVHRVGLPAPGGRRELAEMCGHIAGLALDRDR
MAV_3015|M.avium_104 LNLDHPVWVDDDRFDLGRHLHRIALPSPGGPKELAEICGHVAGLPLDRDH
MSMEG_4297|M.smegmatis_MC2_155 FNLDHPVWVEDNDFDVDRHLHRIGLPGPGGRAELAEICGHIASLPLDRSR
Mvan_3601|M.vanbaalenii_PYR-1 FNLDHPVWVEDNDFDVDRHLHRIGLPSPGGRVELAEICGHIASLPLDRTR
TH_2477|M.thermoresistible__bu FNLDHPVWVEDADFDVDRHLHRIGLPAPGGPAELAEICGHLAALPLDRSR
MAB_1917|M.abscessus_ATCC_1997 LNLDHPVWVDDDDFDIDRHLHHVGLPAPGGKAEIADMCGHIASLPLDRAR
Mflv_5212|M.gilvum_PYR-GCK LNFDHPVWVEDDDFDLTRHLNRVALPEPGGRDELAELCGRLIGMPLERSK
MUL_1818|M.ulcerans_Agy99 LGLDRPWFVEDQDLDVDFHVRRIGVPAPGGRRELEELVGRLMSYKLDRSR
MLBr_01244|M.leprae_Br4923 LPTAAPRWVVDPDFELDFHVRRVRVPDPGTLREVFDLAEVIQQSPMDVSR
: * :* * ::: *:.:: :* ** *: :: : :: :
Rv1760|M.tuberculosis_H37Rv PLWEMWVIEGGARSD-----TVAVMLKVHHAVVDGVAGANLLSHLCSLQP
Mb1791|M.bovis_AF2122/97 PLWEMWVIEGGARSD-----TVAVMLKVHHAVVDGVAGANLLSHLCSLQP
MMAR_2634|M.marinum_M PLWEMWVIEGGARND-----SVTVMLKVHHAVVDGVAGANLLAHLCSFQA
MAV_3015|M.avium_104 PLWEMWVVEGRHGND-----TLSVVLKAHHAVVDGVGGANLLAQLCGTAP
MSMEG_4297|M.smegmatis_MC2_155 PLWEMWVIENVAGTDAHAGGRLALMTKVHHASVDGVTGANLLSQLCTTEP
Mvan_3601|M.vanbaalenii_PYR-1 PLWEMWVIENVAGTDAHAGGRIAVMTKVHHAAVDGVTGANLMSKLCTTEP
TH_2477|M.thermoresistible__bu PLWEMWVIENIDGTDPRSGGPLVVLTKVHHAAVDGVTGANLLSQVCTTEA
MAB_1917|M.abscessus_ATCC_1997 PLWEMWVIETGDTN------RLVVMTKMHHASVDGVTGANLMSALCGLEP
Mflv_5212|M.gilvum_PYR-GCK PLWEMWVIEGVGGSAAREDGRLALLLKVHHAAVDGVSAADLLAKLLDTEP
MUL_1818|M.ulcerans_Agy99 PLWELWVIEGVEGG------RIATLTKMHHAIVDGVSGAGLGEILLDVTP
MLBr_01244|M.leprae_Br4923 PLWTATLVEGLAAG------RAAMLLQISHAITDGVGSVEMFAEMYDLER
*** ::* : : ** .*** .. : :
Rv1760|M.tuberculosis_H37Rv DAPAP--QPVRGTGGGN----------VLQIAASGLEG--FASRPVRLAT
Mb1791|M.bovis_AF2122/97 DAPAP--QPVRGTGGGN----------VLQIAASGLVG--FASRPVRLAT
MMAR_2634|M.marinum_M DGPVP--LPARGAGPGH----------PLQIATSGLMG--FALRPLRLAT
MAV_3015|M.avium_104 DAPPP--EPAPRTGTVN----------RLQIAAGGLIG--AALRPWRLAR
MSMEG_4297|M.smegmatis_MC2_155 DAPPP--DPVDGPGGGT----------GLEIAVNGAVR--FATRPLRLVN
Mvan_3601|M.vanbaalenii_PYR-1 DAPPP--DPVAGPGGGT----------DLEIAVSGAVR--FATRPLKLVN
TH_2477|M.thermoresistible__bu DTPPP--EPVQGAGTAS----------DLEIALSGAVR--FATRPLKLLN
MAB_1917|M.abscessus_ATCC_1997 DAEAP--EPAPGVGGAN----------SLEIAVTGALK--WASRPLKFAK
Mflv_5212|M.gilvum_PYR-GCK DAPPP--DPVDAPTGAS----------TWEIAADGLWR--VATRPWQLTR
MUL_1818|M.ulcerans_Agy99 EPRPPQEETVGFVGFQIP---------RLERRAVGALINVGIMTPFRIAR
MLBr_01244|M.leprae_Br4923 DPPSRPRSPQPIPQDLSRNDLMLQGINHLPVALAGGVVGGLSGVASVAGR
: . * .
Rv1760|M.tuberculosis_H37Rv VVPATVLTLVRTLLRAREG-RTMAAPFSAPPTPFNGPLGRLRNIAYTQLD
Mb1791|M.bovis_AF2122/97 VVPATVLTLVRTLLRAREG-RTMAAPFSAPPTPFNGPLGRLRNIAYTQLD
MMAR_2634|M.marinum_M VVPATIRTLAQTVLRAREG-RTMAAPFSAPPTPFNGNVTRHRNVAFTQLD
MAV_3015|M.avium_104 VLPATALTLAETVLRARGGGQTMAAPFAAPATPFNGTFTRRRNVALAGVD
MSMEG_4297|M.smegmatis_MC2_155 VVPSTLNTVVDTVRRARNG-RTMAAPFAAPRTAFNANITGHRNIAFAQLD
Mvan_3601|M.vanbaalenii_PYR-1 VVPNTVSTVIDTVKRARNG-LTMAPPFAAPKTAFNANVTGHRNVAFAQLD
TH_2477|M.thermoresistible__bu VLPTTANSVLDTVRRARAG-RTMARPFAAPSTPFNANVTGRRNVAFTQLD
MAB_1917|M.abscessus_ATCC_1997 LLPATIGVIPAWLERSKRG-EAMSAPFSAPRTSFNSTITSRRNVGYAQLD
Mflv_5212|M.gilvum_PYR-GCK VVPVATSLVTNTVSRALSG-TAMALPFSAPSTPFNAPLTSDRNIAFAQLD
MUL_1818|M.ulcerans_Agy99 LVEQTVRQQIAALGVSARP----PRYFDAPKTRFNAHVSPHRRVSGCRIE
MLBr_01244|M.leprae_Br4923 AILRPASTVSGVVGYVRSG--IRVLSQAAEPSPLLRQRSLATRTEAIEIQ
: . : * : : . ::
Rv1760|M.tuberculosis_H37Rv MRDVKRVKDRFGVTINDVVVALCAGALRRFLLEHGVLPEAPLVATVPVSV
Mb1791|M.bovis_AF2122/97 MRDVKRVKDRFGVTINDVVVALCAGALRRFLLEHGVLPEAPLVATVPVSV
MMAR_2634|M.marinum_M MRDVKRVKQRFGVTVNDVVVALCAGVLRRFLLERGELPDTPLVATVPVSV
MAV_3015|M.avium_104 LEDVKAVKRHFGVTVNDVVTAICAGALRQFLADRDALPEVPLVASVPVSV
MSMEG_4297|M.smegmatis_MC2_155 LEDIKTVKNHFDVKVNDVVMALVSGVLRTFLSHRDELPDNSLVAMVPVSV
Mvan_3601|M.vanbaalenii_PYR-1 LEDIKTVKNHFGVKVNDVVMALVSGVLRTFLLERGELPESSLVAMVPVSV
TH_2477|M.thermoresistible__bu LEDVKRIKDHFGVKVNDVVMALTSGALRSYLLERDQLPDSPLIAMVPVSV
MAB_1917|M.abscessus_ATCC_1997 LEDVRTVKNHFGVKVNDVVMAICAGALRKYLDDRGELPDNSLVAMVPVSV
Mflv_5212|M.gilvum_PYR-GCK LDDVKKVKNQAEVKVNDVVMALCAGALRGYLDERGALPDKPLIAVVPSSV
MUL_1818|M.ulcerans_Agy99 LARAKAVKDAFGVKLNDVVLATVAGAAREYLKKRDELPAKPLIAQIPVST
MLBr_01244|M.leprae_Br4923 LSDLHKAAKAGDGSINDAYLAGLVGALRRYHEALG-VSISTLPMAVPVNV
: : .:**. * *. * : . :. .* :* ..
Rv1760|M.tuberculosis_H37Rv HDKSDRPGR-NQATWMFCRVPSQISDPAQRIRTIAAGNTVAKDHAAAIGP
Mb1791|M.bovis_AF2122/97 HDKSDRPGR-NQATWMFCRVPSQISDPAQRIRTIAAGNTVAKDHAAAIGP
MMAR_2634|M.marinum_M HEKSDRPGR-NQTTWMFCRVASQISDPAERIRAIAKGNAAAKDHTAAVGP
MAV_3015|M.avium_104 HGKSSRPGR-NQTTWMLCRLETHIDDPVHRLTTIAEKNAAAKEHAAAMGP
MSMEG_4297|M.smegmatis_MC2_155 HDRSDRPGR-NQVSGMFSPLQTHIADPAERLRAIAEANAVAKQHSSAIGA
Mvan_3601|M.vanbaalenii_PYR-1 HDKSDRPGR-NQVSGMFSRLETNIEDPATRLKAIAEANSVAKQHSSAIAA
TH_2477|M.thermoresistible__bu HDKTDRPGR-NHVSGMFSRLETTIEDPAERLRAIANANSVAKQHSSAIAA
MAB_1917|M.abscessus_ATCC_1997 HEKSDRPGR-NQVSGMFSRLETNVDDPVERLNAIAAANNIAKDHTAVLGA
Mflv_5212|M.gilvum_PYR-GCK HGQTDRPGR-NQLSGMFCNLHTDIEDPLDRLYAIGASNVRAKEHSGSLGP
MUL_1818|M.ulcerans_Agy99 RTDQTKEDVGNQISSMTASLATHINDPAERLKAIHESTQNAKEMAKALSA
MLBr_01244|M.leprae_Br4923 RTEADVVGS-NRFVGVTLAAPLGTNDPAARMQKIRSQMTQRRDEPAMNII
: . *: : ** *: * :: .
Rv1760|M.tuberculosis_H37Rv TLLHDWIQFGGSTMFGAAMRILPHISITHSPA--YNLILSNVPGPQAQLY
Mb1791|M.bovis_AF2122/97 TLLHDWIQFGGSTMFGAAMRILPHISITHSPA--YNLILSNVPGPQAQLY
MMAR_2634|M.marinum_M TLLHDWTQFGGPTMF----SILPRIPISRSPA--YNLILSNVPGPQTQLY
MAV_3015|M.avium_104 TLLQDWTQVAGQTVFGVARKLLPRIPLPDKPP--HNLVLSNVAGPRHQLY
MSMEG_4297|M.smegmatis_MC2_155 TLLQDWTQFAAPAVFGTAMRVYAASRLSGAKP-VHNLVVSNVPGPQVPLY
Mvan_3601|M.vanbaalenii_PYR-1 TLLQDWTQFAAPAVFGAAMRVYASSRLSGARP-VHNLVISNVPGPQGPLY
TH_2477|M.thermoresistible__bu SLLQDWTQFAAPAVFGVAMRVYAASRLGEAKP-VHNLVVSNVPGPQVPLY
MAB_1917|M.abscessus_ATCC_1997 TLLQDWSQFAAPAVFGTAMRVYSRIRLADRHPVIHNLVVSNVPGPQVPLY
Mflv_5212|M.gilvum_PYR-GCK SMLMDLAQSVSRTMFGWLLGGLSRTSLGNTAI--HNVIISNVAGPPTTLY
MUL_1818|M.ulcerans_Agy99 HQIMGLTETTPPGLLHLAARAYTATGLSHNLA-PINLVVSNVPGPQFPLY
MLBr_01244|M.leprae_Br4923 GSLAPLMTVLPASVLDFIVDSVASSDVN----------ASNIPAYPGDTY
: :: . : **:.. *
Rv1760|M.tuberculosis_H37Rv FLGCRMDSMFPLGPLLGNAGLNITVMS-----LNGELGVGIVSCPDLLP-
Mb1791|M.bovis_AF2122/97 FLGCRMDSMFPLGPSLATRASTSPSCP-----STGNWVSALSPAPTCCR-
MMAR_2634|M.marinum_M FLGCRVDAMCPLGPILGGAGLNITVMS-----LNGELGIGLISCPDLVP-
MAV_3015|M.avium_104 FLGCRLDGMYPLGPIIAGAALNITVMS-----LGGRLGVGILSCPDLVP-
MSMEG_4297|M.smegmatis_MC2_155 FLGAEVKAMYPLGPIFHGSGLNITVMS-----LTGKLDVGIISCPELLP-
Mvan_3601|M.vanbaalenii_PYR-1 MLGCEVKAMYPLGPIFHGSGLNMTVMS-----LSGKLDVGIVSCPELLP-
TH_2477|M.thermoresistible__bu FLGAEVKAMYPLGPIFHGSGLNITVMS-----LTGRLNVGMVSCPELMP-
MAB_1917|M.abscessus_ATCC_1997 FLGAQVIGMYPLGPIFHGAALTVTVMS-----LDGKLNVGLISCPDLMP-
Mflv_5212|M.gilvum_PYR-GCK SGGAEITGLYPLGPIFPGSGLNITVMS-----MGDKLNVGIISCPKLTD-
MUL_1818|M.ulcerans_Agy99 MAGARLDSLVPLGPPVMDVALNITCFS-----YQEHLDFGFVTTPEVAN-
MLBr_01244|M.leprae_Br4923 FAGAKILRQYGIGPRPGVAMMAVLMSRGGFCTVTVRYDRASVKSEALFAR
*..: :** . .
Rv1760|M.tuberculosis_H37Rv -DLWGVADGFP-------EALKELLECSDDQPEGSNHQDS
Mb1791|M.bovis_AF2122/97 -TCGAWQTGFPRRSKSCWSAVMTSRKAATTRTPESYVQNR
MMAR_2634|M.marinum_M -DLWGLADGFP-------DALKELLECRDVRCADIDLVDV
MAV_3015|M.avium_104 -DLWDLADAFP-------AALKELTNCVRPGRART-----
MSMEG_4297|M.smegmatis_MC2_155 -DLWDMADDFA-------VALDELLDATKRA---------
Mvan_3601|M.vanbaalenii_PYR-1 -DLWDMADTFG-------VALQELLDATR-----------
TH_2477|M.thermoresistible__bu -DLWELAEGFE-------RALRELLDTVG-----------
MAB_1917|M.abscessus_ATCC_1997 -DLGTLTDDFG-------VALEELKARI------------
Mflv_5212|M.gilvum_PYR-GCK -DLWDLADRFD-------AELADLLARS------------
MUL_1818|M.ulcerans_Agy99 -DIDEMADLIEP-------ALAELERAAGL----------
MLBr_01244|M.leprae_Br4923 CLLEGFDEVLALAGDPTPHAVPASFAARSSGSPAGWLSSS
: :