For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AARKCGAPPIAADGSTRRPDCVTAVRTQARAPTQHYAESVARRQRVLTITAWLAVVVTGSFALMQLATGA GGWYIALINVFTAVTFAIVPLLHRFGGLVAPLTFIGTAYVAIFAIGWDVGTDAGAQFFFLVAAALVVLLV GIEHTALAVGLAAVAAGLVIALEFLVPPDTGLQPPWAMSVSFVLTTVSACGVAVATVWFALRDTARAEAV MEAEHDRSEALLANMLPASIAERLKEPERNIIADKYDEASVLFADIVGFTERASSTAPADLVRFLDRLYS AFDELVDQHGLEKIKVSGDSYMVVSGVPRPRPDHTQALADFALDMTNVAAQLKDPRGNPVPLRVGLATGP VVAGVVGSRRFFYDVWGDAVNVASRMESTDSVGQIQVPDEVYERLKDDFVLRERGHINVKGKGVMRTWYL IGRKVAADPGEVRGAEPRTAGV*
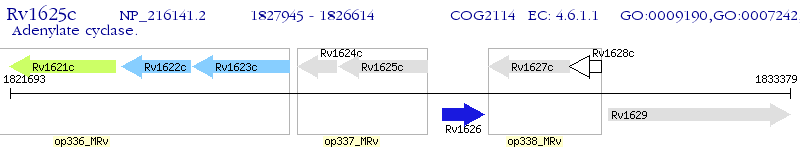
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1625c | cya | - | 100% (443) | MEMBRANE-ANCHORED ADENYLYL CYCLASE CYA (ATP PYROPHOSPHATE-LYASE) (ADENYLATE CYCLASE) |
| M. tuberculosis H37Rv | Rv2435c | - | 5e-31 | 33.81% (210) | cyclase |
| M. tuberculosis H37Rv | Rv1318c | - | 2e-13 | 34.25% (146) | adenylate cyclase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1651c | cya | 0.0 | 100.00% (418) | membrane-anchored adenylyl cyclase Cya (ATP |
| M. gilvum PYR-GCK | Mflv_3576 | - | 1e-132 | 58.51% (388) | putative adenylate/guanylate cyclase |
| M. leprae Br4923 | MLBr_02341 | - | 3e-15 | 29.24% (171) | putative regulatory protein |
| M. abscessus ATCC 19977 | MAB_0490c | - | 9e-13 | 28.12% (192) | putative adenylate cyclase |
| M. marinum M | MMAR_2428 | cya | 1e-180 | 72.85% (442) | membrane-anchored adenylyl cyclase Cya |
| M. avium 104 | MAV_3162 | - | 0.0 | 72.01% (443) | adenylate cyclase |
| M. smegmatis MC2 155 | MSMEG_3578 | - | 1e-24 | 25.74% (272) | putative cyclase |
| M. thermoresistible (build 8) | TH_2942 | - | 1e-135 | 56.93% (411) | PUTATIVE adenylate cyclase |
| M. ulcerans Agy99 | MUL_3698 | - | 1e-24 | 27.94% (204) | adenylate or guanylate cyclase |
| M. vanbaalenii PYR-1 | Mvan_2839 | - | 1e-136 | 57.77% (412) | putative adenylate/guanylate cyclase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3576|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2839|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2942|M.thermoresistible__bu --------------------------------------------------
Rv1625c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1651c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2428|M.marinum_M --------------------------------------------------
MAV_3162|M.avium_104 --------------------------------------------------
MSMEG_3578|M.smegmatis_MC2_155 -------------------MLSRISIQSKLLAMLLVTSVLSAAVVGFIGY
MUL_3698|M.ulcerans_Agy99 -----------------MLRFLRVGIPAKLLVTLLVCSILSVAVVGVIGY
MAB_0490c|M.abscessus_ATCC_199 -----MTGAEGRPDRRPLARAARYSRWVMRTPWPLFIMGIAQSNLVGALF
MLBr_02341|M.leprae_Br4923 MTTALVCVACGAELRSIAKFCDECGSPVVEAETPAEYKQVTVLFADVVGS
Mflv_3576|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2839|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2942|M.thermoresistible__bu --------------------------------------------------
Rv1625c|M.tuberculosis_H37Rv ----------------------------------------MAARKCGAPP
Mb1651c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2428|M.marinum_M ------------------------------------MPAKKSAKKCRTAR
MAV_3162|M.avium_104 ----------------------------------------MAAKSCGAAP
MSMEG_3578|M.smegmatis_MC2_155 QSGRSSLRDSVFDRLTEIRTAQARQLETQFRSFRNSLVIYTRGTTAIEAA
MUL_3698|M.ulcerans_Agy99 VSGRNALQRVAVERLHQLREAQKRQVEALFSELTKSVMVYSNGMSANEAT
MAB_0490c|M.abscessus_ATCC_199 VLGFLRFGLPPSDRVELTELGPTRLIILGFSLFILFVVGAIVSTYLLLPV
MLBr_02341|M.leprae_Br4923 MDIAAAVGLERLREIMTELVSRAAGVIERYGGTVDKFTGDGIMAIFGAPT
Mflv_3576|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2839|M.vanbaalenii_PYR-1 -----MRPVCVPEVTLGRRAWSPSRHRHER--------------------
TH_2942|M.thermoresistible__bu --------------VARQGVPARTQRYAES--------------------
Rv1625c|M.tuberculosis_H37Rv IAADGSTRRPDCVTAVRTQARAPTQHYAES--------------------
Mb1651c|M.bovis_AF2122/97 ---------------MRTQARAPTQHYAES--------------------
MMAR_2428|M.marinum_M RSSDG-LPRPDSVAAVREQTRDLNQHYAHS--------------------
MAV_3162|M.avium_104 LWPTGSSKRPDCVAAARAQSRARNQHYADS--------------------
MSMEG_3578|M.smegmatis_MC2_155 RAFTAGFDQLGDATISPAQQKAITDWYENQFSATTEHSDAADNLDGNEVD
MUL_3698|M.ulcerans_Agy99 AALTDGFAQLANATVDPVQQQAIVDYYVN------ELIKPIDQATGHELD
MAB_0490c|M.abscessus_ATCC_199 LRWQYRSPDFESEGSEAVRVRALQMPFYRT--------------------
MLBr_02341|M.leprae_Br4923 ALEDHALRACLAALGVQEKARRLAAEVGDRDGVDLQLRVGLNSGQVIVGE
Mflv_3576|M.gilvum_PYR-GCK -------MGRRQSVLMIAGWIAAAVSAAFGAFQLTLGGSLWWLGAVNVLC
Mvan_2839|M.vanbaalenii_PYR-1 -------VARRQRVLMIASWIAAAASVGFGVFQLTFGGPLWWLGAVNIGC
TH_2942|M.thermoresistible__bu -------IRSRYRMLTIAAWIAAAVSAGFGTLQLVLGGQTRLLGVINLAV
Rv1625c|M.tuberculosis_H37Rv -------VARRQRVLTITAWLAVVVTGSFALMQLATGAGGWYIALINVFT
Mb1651c|M.bovis_AF2122/97 -------VARRQRVLTITAWLAVVVTGSFALMQLATGAGGWYIALINVFT
MMAR_2428|M.marinum_M -------AARRARVLSIAALLAVLISSSFILVQFLTQAWIWQITLINIAA
MAV_3162|M.avium_104 -------AARQSRVLAIAAWLAVLVCADFILLQVVTGAWTWQIVSLNVVA
MSMEG_3578|M.smegmatis_MC2_155 V---EALLPSSNPQKYLQALYTAPFTDWDKAIENDDAGDGSGWSAANALF
MUL_3698|M.ulcerans_Agy99 L---EAVLSASPAQTYLQAYYTAPFSSSSESMRFNNAGDGSAWSAANARF
MAB_0490c|M.abscessus_ATCC_199 ------VISVATWLVGGAIFVAVTWSSTHSSVLVAAFATVLGATTTSIIG
MLBr_02341|M.leprae_Br4923 IGSSALGYTAVGVQVGMAQRMESVAPPGVVMLSESTARLVEQTAVLGAPQ
. .
Mflv_3576|M.gilvum_PYR-GCK ALVFLWIPRLCPLGELLAPLTFVTFAYMS---------------------
Mvan_2839|M.vanbaalenii_PYR-1 GVVFLAIPRLCPLGELIAPLTFVVFAYTS---------------------
TH_2942|M.thermoresistible__bu AVVFTLIPLLYRFGELLAPLSFFLFAYLS---------------------
Rv1625c|M.tuberculosis_H37Rv AVTFAIVPLLHRFGGLVAPLTFIGTAYVA---------------------
Mb1651c|M.bovis_AF2122/97 AVTFAIVPLLHRFGGLVAPLTFIGTAYVA---------------------
MMAR_2428|M.marinum_M TLVFATVPMLHRFGELVAPLTFVGAAYVS---------------------
MAV_3162|M.avium_104 ALIFAIVPWLHRFGDLVAPLTFLGAAYVT---------------------
MSMEG_3578|M.smegmatis_MC2_155 NGFFREIVTRSGFDDALLLDTRGNVVYTAYKGSDLGTNIYTGPYKGGELS
MUL_3698|M.ulcerans_Agy99 NDFFRGIVTRFQYRDALLLDLQGNVVYSVNEGPDLGTNLFTGPYRETNLR
MAB_0490c|M.abscessus_ATCC_199 YLQSERVLRPVAIQALRRGVPENVTAPGVIVR------------------
MLBr_02341|M.leprae_Br4923 TVRIKGAEDAVPARRLLRVAEPRQPTWFSESTLVG---------------
.
Mflv_3576|M.gilvum_PYR-GCK ------------------------------VTFFCYTIGTGSGMQ--FYF
Mvan_2839|M.vanbaalenii_PYR-1 ------------------------------VGFICYTIGTGSGLQ--FYF
TH_2942|M.thermoresistible__bu ------------------------------VGFMCFSIGTGSGLQ--LYL
Rv1625c|M.tuberculosis_H37Rv ------------------------------IFAIGWDVGTDAGAQ--FFF
Mb1651c|M.bovis_AF2122/97 ------------------------------IFAIGWDVGTDAGAQ--FFF
MMAR_2428|M.marinum_M ------------------------------VFASCWNVGTGSGAQ--MFF
MAV_3162|M.avium_104 ------------------------------VFVSCWDVGTASGGQ--YFF
MSMEG_3578|M.smegmatis_MC2_155 EAYREALNSNTVDYVAVTDFGIYQPS-GTPTAWMVSPIGGGGRIEGVLAL
MUL_3698|M.ulcerans_Agy99 TAYQKALASNEIDFVWITDFQRYQPEVDAPTAWLVSPVGVDGKLQGVMAL
MAB_0490c|M.abscessus_ATCC_199 -------------------------------QILTWALSTGVPILAIVLT
MLBr_02341|M.leprae_Br4923 -------REWELATLAAMLDRSIGGRGSVVGLVGPAGIGKTRLVAEAVQL
:.
Mflv_3576|M.gilvum_PYR-GCK LVAASLVLLVLGIERVALAAVIAGVGVVITVVLELTVPEDRGLGP-----
Mvan_2839|M.vanbaalenii_PYR-1 LVAASLVVLVLGIERIALAAAIAAAGVIITIVLELTVPDDRGLGP-----
TH_2942|M.thermoresistible__bu LVAASLVVLLLGVDRIVLAAGIVAVGVVGIIVLELVVPEDRGLGP-----
Rv1625c|M.tuberculosis_H37Rv LVAAALVVLLVGIEHTALAVGLAAVAAGLVIALEFLVPPDTGLQP-----
Mb1651c|M.bovis_AF2122/97 LVAAALVVLLVGIEHTALAVGLAAVAAGLVIALEFLVPPDTGLQP-----
MMAR_2428|M.marinum_M LVGACLVVLVLGIEHVVLASGLAALAAALIITVEFVVPRNTGAQS-----
MAV_3162|M.avium_104 LVGACLVLLVMGIEHTVLAAVLAAIGAGLVIAVEFAFPRDTGLQP-----
MSMEG_3578|M.smegmatis_MC2_155 QFPISVVNQLMTMDGKWEESGLGETGETYLVGPDGLMRSDSRLFLENPEQ
MUL_3698|M.ulcerans_Agy99 ALPPTNLNQIMNANEKWEAVGMGRSTETYLAGSDNLMRSDSRLFLESPDE
MAB_0490c|M.abscessus_ATCC_199 VVGQRAGYLKSNADNLTGSILILAIIALAIGLMGTLLVATSIADP-----
MLBr_02341|M.leprae_Br4923 AKGLEVEVFSVFCESHATDVAFDVVAQLVRAVAQISGLDDRSARARVREQ
: :
Mflv_3576|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2839|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_2942|M.thermoresistible__bu --------------------------------------------------
Rv1625c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1651c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2428|M.marinum_M --------------------------------------------------
MAV_3162|M.avium_104 --------------------------------------------------
MSMEG_3578|M.smegmatis_MC2_155 FKADVIEAGTPPGVAEQSILHGGTTLVQPVSDEASKLAGQGQSGSIIERD
MUL_3698|M.ulcerans_Agy99 YRREALAGGTREDVVNKAMWLGTTVLVQPVPSAGLRAALRGQSGITKDTD
MAB_0490c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_02341|M.leprae_Br4923 IPDADTQDLLLLDDLLG-------------------------IADPAVEL
Mflv_3576|M.gilvum_PYR-GCK ----------------------------------------SWTLTAGFIS
Mvan_2839|M.vanbaalenii_PYR-1 ----------------------------------------SWTLTAGFIS
TH_2942|M.thermoresistible__bu ----------------------------------------AWMLTAGFVT
Rv1625c|M.tuberculosis_H37Rv ----------------------------------------PWAMSVSFVL
Mb1651c|M.bovis_AF2122/97 ----------------------------------------PWAMSVSFVL
MMAR_2428|M.marinum_M ----------------------------------------AWLQSLGFII
MAV_3162|M.avium_104 ----------------------------------------AWAQSMSFVV
MSMEG_3578|M.smegmatis_MC2_155 YLGQETLQAYAPLDLTG--LDWSVVAKIDTAEAFAPVTKFTRTLVLSTVA
MUL_3698|M.ulcerans_Agy99 YLGNRELEAYAPLDVPNSDLRWAILATRDDWDAYERLSSFGRTVVLAVAG
MAB_0490c|M.abscessus_ATCC_199 ------------------------------------------LRQLRWAL
MLBr_02341|M.leprae_Br4923 PRLDPDARRRRLTALINVAQLALSRPAVFVVEDVHWIDEVSDSMLADFLA
Mflv_3576|M.gilvum_PYR-GCK AVTSSAVMIVATIWYTLREIERAES-------------------------
Mvan_2839|M.vanbaalenii_PYR-1 AVVSSVVIIVATIWYTLREIDRAEA-------------------------
TH_2942|M.thermoresistible__bu SAVSSALIIFATLWYALREIARTEA-------------------------
Rv1625c|M.tuberculosis_H37Rv TTVSACGVAVATVWFALRDTARAEA-------------------------
Mb1651c|M.bovis_AF2122/97 TTVSACGVAVATVWFALRDTARAEA-------------------------
MMAR_2428|M.marinum_M TTVSACLLVFVTVWFALRDTARAEA-------------------------
MAV_3162|M.avium_104 TIVSACIMVFVAVWFALRDTQRAEA-------------------------
MSMEG_3578|M.smegmatis_MC2_155 MIFAVCIAAILLARVFVRPIRRLEAGAQQISAGDYGVALPVVSRDEFGDL
MUL_3698|M.ulcerans_Agy99 ITLFICVVSMVLAKLLVRPIRRLEAGTEKIRSGDYEVTVPVKSRDELGDL
MAB_0490c|M.abscessus_ATCC_199 SEVQRGNYNAHVPIYDASELGLLQAGFN----------------------
MLBr_02341|M.leprae_Br4923 VIPQTRSMALVTYRPEYHGALRHVVGAQIIAVAPLSDVETSTLVAELLG-
Mflv_3576|M.gilvum_PYR-GCK ----------------AMENEYERSERLLANILPATIAER---LKEPTRT
Mvan_2839|M.vanbaalenii_PYR-1 ----------------AMAAEYERSEELLANILPATIADR---LKEPTRT
TH_2942|M.thermoresistible__bu ----------------AMEAEYQRSESLLLNILPASVARR---LKDPART
Rv1625c|M.tuberculosis_H37Rv ----------------VMEAEHDRSEALLANMLPASIAER---LKEPERN
Mb1651c|M.bovis_AF2122/97 ----------------VMEAEHDRSEALLANMLPASIAER---LKEPERN
MMAR_2428|M.marinum_M ----------------VMEAQYDRSEALLANMLPVSVADR---LKEPERN
MAV_3162|M.avium_104 ----------------VMESEYQRSEALLANMLPGSIAER---LKHSDGD
MSMEG_3578|M.smegmatis_MC2_155 TVAFNEMSRNLRIKEDLLDEQRKENDRLLLSLMPEPVVQR---YRE-GEE
MUL_3698|M.ulcerans_Agy99 TAAFNEMSSNLKVKDQLLTEQRSQNEQLLLALMPEPVVQR---YRA-DGE
MAB_0490c|M.abscessus_ATCC_199 -------------DMVRDLGERQRLRDLFGRYVGEDVARR---ALEYGTE
MLBr_02341|M.leprae_Br4923 -------LDQSVTQIGELITERAAGNPFFAQEITRELAERGVLTGERGRY
: :: : :. *
Mflv_3576|M.gilvum_PYR-GCK VIADKYDDASILFADIAGYTKRASDTAPAELVRFLDLLYTDLDALVDRHG
Mvan_2839|M.vanbaalenii_PYR-1 VIADKYDDASILFADIAGYTKRASETPPSELVRFLDRLYTDLDALVDRHG
TH_2942|M.thermoresistible__bu VIADKYDDASVLFADIAGYTRWAGATTPSELVLFLDRLYTELDALVDRHG
Rv1625c|M.tuberculosis_H37Rv IIADKYDEASVLFADIVGFTERASSTAPADLVRFLDRLYSAFDELVDQHG
Mb1651c|M.bovis_AF2122/97 IIADKYDEASVLFADIVGFTERASSTAPADLVRFLDRLYSAFDELVDQHG
MMAR_2428|M.marinum_M IIADKYEEASVLFADIVGFTERASSTAPADLVRFLDRLYSAFDALVDKYG
MAV_3162|M.avium_104 IIADKYDDASVLFADIVGFTERASGTAPADLVRFLNRLYIAFDELVDKHG
MSMEG_3578|M.smegmatis_MC2_155 TIAQDHQNVTVLFADLMGLDRLSASMSSDEHLSIVNKLVRQFDAAAESLG
MUL_3698|M.ulcerans_Agy99 VIAEEHQDVTVIFAELIGLEEISDGLSGDELVGIVDELFRQFDSAAQSLG
MAB_0490c|M.abscessus_ATCC_199 LGGQE-RSVAVLFVDLVGSTQLASTRGPAVVVRVLNEFFRVIVDTVNKHG
MLBr_02341|M.leprae_Br4923 ACSTDVAEVCVPATLQATIAARIDRLSPPAKQLLVAAAVIGFRFGSDLLA
. . .. : . .: : : .
Mflv_3576|M.gilvum_PYR-GCK LEKVKTSGDSYMVVSGVPTPRPDHLEALATLALDMSEAVADLKDPRGRDV
Mvan_2839|M.vanbaalenii_PYR-1 LEKVKTSGDSYMVVSGVPNPRADHLEALALLALDMADAVADLKDPQGHDV
TH_2942|M.thermoresistible__bu LEKVKTSGDSYMVVSGVPQPRPDHLEALADFALDMAAAVGRLRDPQGRTV
Rv1625c|M.tuberculosis_H37Rv LEKIKVSGDSYMVVSGVPRPRPDHTQALADFALDMTNVAAQLKDPRGNPV
Mb1651c|M.bovis_AF2122/97 LEKIKVSGDSYMVVSGVPRPRPDHTQALADFALDMTNVAAQLKDPRGNPV
MMAR_2428|M.marinum_M LEKIKVSGDSYMVVSGVPRPRPDHVQALADFALDMVDVAAGMKDPHNRSV
MAV_3162|M.avium_104 LEKIKVSGDSYMVVSGVPRVRPDHATALADFALDMVKVAAALKDPYGRPV
MSMEG_3578|M.smegmatis_MC2_155 VERVRTLHNGYLASCGLTVPRLDNVRRTVDFAFEMQRIVDRFRGETGYDL
MUL_3698|M.ulcerans_Agy99 VEPIRTFHNGYLAACGATTPRLDNVHRGIEFALEIELIVNRFANQSQHPL
MAB_0490c|M.abscessus_ATCC_199 GFVNKFQGDAALAIFGAPIDHPDGAAAALAAARELSGELTNVLGSND---
MLBr_02341|M.leprae_Br4923 SLGIDLSVDELIDANLVDQVR---FTAPVEYAFRHPLIRTVAYESQLKSD
: : : *
Mflv_3576|M.gilvum_PYR-GCK PLRIGMAMGPVVAGVVGAKK-FFYDVWGDAVNVASRMETTDEE---GRIQ
Mvan_2839|M.vanbaalenii_PYR-1 PLRIGMASGPVVAGVVGAKK-FFYDVWGDAVNVASRMETTDVE---GRIQ
TH_2942|M.thermoresistible__bu PLRIGLAAGPVVAGVVGARK-FFYDVWGDAVNVGSRMESTDVV---GRIQ
Rv1625c|M.tuberculosis_H37Rv PLRVGLATGPVVAGVVGSRR-FFYDVWGDAVNVASRMESTDSV---GQIQ
Mb1651c|M.bovis_AF2122/97 PLRVGLATGPVVAGVVGSRR-FFYDVWGDAVNVASRMESTDSV---GQIQ
MMAR_2428|M.marinum_M PLRVGLATGPVVAGVVGSRR-FFYDVWGDAVNVASRMESTDSV---GKIQ
MAV_3162|M.avium_104 PLRVGMACGPVVAGVVGSRR-FFYDVWGDAVNVASRMESTDSV---GRIQ
MSMEG_3578|M.smegmatis_MC2_155 RLRAGIDTGTVSSGLVGRSS-LAYDMWGAAVDLAYQVQRGMPQ---PGVY
MUL_3698|M.ulcerans_Agy99 RVRVGVTTGRVVSGRVGRSN-LAYDMWGGAVQMAYRMLGHLSD---PGIY
MAB_0490c|M.abscessus_ATCC_199 -FGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAARLTELAKNEP-GNVL
MLBr_02341|M.leprae_Br4923 RARLHRRLAAAIEARAPHSVDQHAVLIAEHLEAAGDLHAAYGWHMRAAAW
. . . : . :: . :
Mflv_3576|M.gilvum_PYR-GCK VPDNVYQRLRHAFVLEERGLIDVKGKGPMHTWYLVGHR------------
Mvan_2839|M.vanbaalenii_PYR-1 VPDNVYERLRDRFVLEERGVVDVKGKGPMHTWYLVAHR------------
TH_2942|M.thermoresistible__bu VAQDVRDRLRDSFVFEERGAIEVKGKGVMHTWYLLGRRSDRQGSDRQGSG
Rv1625c|M.tuberculosis_H37Rv VPDEVYERLKDDFVLRERGHINVKGKGVMRTWYLIGRKVAADPGE-----
Mb1651c|M.bovis_AF2122/97 VPDEVYERLKDDFVLRERGHINVKGKGVMRTWYLIGRKVAADPGE-----
MMAR_2428|M.marinum_M VPDEVYERLNKDFVLQERGHINVKGKGVMRTWYLIGRKPIDEPTA-----
MAV_3162|M.avium_104 VPETMYERLASEFVLQERGRIEVKGKGVMRTWYLIGRKPAEAPTD-----
MSMEG_3578|M.smegmatis_MC2_155 VTDQVHDAVRDIRQFTAVGSVTVDGTE-RPIWRLSERQS-----------
MUL_3698|M.ulcerans_Agy99 VSARVHDAMREIWTFTPAGAISVRGSQ-QQIWQVSERR------------
MAB_0490c|M.abscessus_ATCC_199 ASASAVMEARDEEALRWDVGETVELRGRTVSTQVARPRHG----------
MLBr_02341|M.leprae_Br4923 ATNQDVRAARVSWEHATKIADALTAEDPHRAAMCIAPRTRLCGIAWRGRF
.. : :
Mflv_3576|M.gilvum_PYR-GCK --------------ARAVTPSSSPP---------------
Mvan_2839|M.vanbaalenii_PYR-1 --------------AEAVTPSGSPT---------------
TH_2942|M.thermoresistible__bu RQGSGRQGSGRQGGTEASTAARTHG---------------
Rv1625c|M.tuberculosis_H37Rv --------------VRGAEPRTAGV---------------
Mb1651c|M.bovis_AF2122/97 --------------VRGAEPRTAGV---------------
MMAR_2428|M.marinum_M --------------VPADEPNAARV---------------
MAV_3162|M.avium_104 --------------LVAEQPHTAHV---------------
MSMEG_3578|M.smegmatis_MC2_155 ----------------------------------------
MUL_3698|M.ulcerans_Agy99 ----------------------------------------
MAB_0490c|M.abscessus_ATCC_199 ----------------------------------------
MLBr_02341|M.leprae_Br4923 NSSSGFDEVRQLCNATGDKAPLAVATVGPAIDHVYQDRLH