For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAKKSTAQRLGRVLETVTRQSGRLPETPAYGSWLLGRVSESQRRRRVRIQVMLTALVVTANLLGIGVALL LVTIAIPEPSIVRDTPRWLTFGVVPGYVLLALALGSYALTRQTVQALRWAIEGRKPTREEERRTFLAPWR VAVGHLMFWGVGTALLTTLYGLINNAFIPRFLFAVSFCGVLVATATYLHTEFALRPFAAQALEAGPPPRR LAPGILGRTMVVWLLGSGVPVVGIALMAMFEMVLLNLTRMQFATGVLIISMVTLVFGFILMWILAWLTAT PVRVVRAALRRVERGELRTNLVVFDGTELGELQRGFNAMVAGLRERERVRDLFGRHVGREVAAAAERERS KLGGEERHVAVVFIDIVGSTQLVTSRPPADVVKLLNKFFAIVVDEVDRHHGLVNKFEGDASLTIFGAPNR LPCPEDKALAAARAIADRLVNEMPECQAGIGVAAGQVIAGNVGARERFEYTVIGEPVNEAARLCELAKSR PGKLLASAQAVDAASEEERARWSLGRHVKLRGHDQPVRLAKPVGLTKPRR*
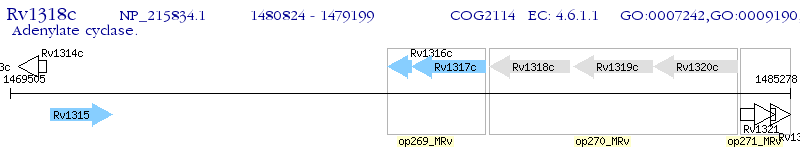
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1318c | - | - | 100% (541) | POSSIBLE ADENYLATE CYCLASE (ATP PYROPHOSPHATE-LYASE) (ADENYLYL CYCLASE) |
| M. tuberculosis H37Rv | Rv1319c | - | 0.0 | 70.97% (534) | adenylate cyclase |
| M. tuberculosis H37Rv | Rv1320c | - | 0.0 | 68.73% (534) | adenylate cyclase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1352c | - | 0.0 | 100.00% (541) | adenylate cyclase |
| M. gilvum PYR-GCK | Mflv_2336 | - | 0.0 | 67.60% (534) | putative adenylate/guanylate cyclase |
| M. leprae Br4923 | MLBr_00201 | - | 2e-93 | 41.46% (451) | hypothetical protein MLBr_00201 |
| M. abscessus ATCC 19977 | MAB_1459c | - | 1e-170 | 55.85% (530) | hypothetical protein MAB_1459c |
| M. marinum M | MMAR_4078 | - | 0.0 | 77.63% (532) | adenylate cyclase |
| M. avium 104 | MAV_1540 | - | 0.0 | 76.45% (535) | HAMP domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4924 | - | 0.0 | 64.86% (535) | adenylate cyclase, family protein 3 |
| M. thermoresistible (build 8) | TH_3474 | - | 0.0 | 68.86% (533) | PUTATIVE putative adenylate/guanylate cyclase |
| M. ulcerans Agy99 | MUL_3944 | - | 0.0 | 67.16% (539) | adenylate cyclase |
| M. vanbaalenii PYR-1 | Mvan_4309 | - | 0.0 | 69.35% (535) | putative adenylate/guanylate cyclase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1318c|M.tuberculosis_H37Rv ----------------MSAKKSTAQRLGRVLETVTRQSGRLPETP-AYGS
Mb1352c|M.bovis_AF2122/97 ----------------MSAKKSTAQRLGRVLETVTRQSGRLPETP-AYGS
MMAR_4078|M.marinum_M MPSTKTAETGSKKGAKSGAKKGVAQRLGRVLERVTRQSGRLPETP-AYGS
MAV_1540|M.avium_104 ----------------MSATQSTAQRIGRVLEKITRQSGRLPETP-AYGS
MUL_3944|M.ulcerans_Agy99 ----------------MVATNTVVQRLRRALEKITQQSDQLTENY-AYGS
Mflv_2336|M.gilvum_PYR-GCK ----------------MNATKSVPYRLGRVLERVTRQSGRLDTP--GYGS
Mvan_4309|M.vanbaalenii_PYR-1 ----------------MNATKSVPYRLGRVLERVTRQSGRLETP--DYGS
TH_3474|M.thermoresistible__bu ----------------MTPNNRLARRLGHLLETLTRQSGRLSGAN-EYGS
MSMEG_4924|M.smegmatis_MC2_155 ----------------MNADNSLARQLGRVLETVTRQNSRLPSGTPEYGS
MAB_1459c|M.abscessus_ATCC_199 --------------MANESIRAVSRRVGRALGAITGQTGNINTGN-GYGS
MLBr_00201|M.leprae_Br4923 ---------------------------------MATQAAGLPGRIGAFVR
:: * : :
Rv1318c|M.tuberculosis_H37Rv WLLGRVSESQRRRRVRIQVMLTALVVTANLLGIGVALLLVTIAIPEPSIV
Mb1352c|M.bovis_AF2122/97 WLLGRVSESQRRRRVRIQVMLTALVVTANLLGIGVALLLVTIAIPEPSIV
MMAR_4078|M.marinum_M WLLGRVSESQQRRRIRIQTILTVMVVVANLLGIAVSILLVTVAIPSPSVF
MAV_1540|M.avium_104 LLLGRVTESQHRRRIRIQIIMTVMVLGANLIGIGAALILVIVAIPAPSVF
MUL_3944|M.ulcerans_Agy99 RLLGQLTETPQRRRVRIQLMLTSSIMAANLVAIGVALLLVFVAIPEPNVI
Mflv_2336|M.gilvum_PYR-GCK WLLGRPAESQSRRRVRIQIILTFFILFTNIMGIAVSLLLNTIAIPVPSVF
Mvan_4309|M.vanbaalenii_PYR-1 WLLGKPEESQARRRIRIQVILTFLLLFTNILGIAVSLLLNTVAIPVPSVF
TH_3474|M.thermoresistible__bu WLLGRVSESQRRRRLRIQTILTTFIVGANVVGIAVAVTLVSFVFPVPSIF
MSMEG_4924|M.smegmatis_MC2_155 WILGRVTESQSRRRIRIQLILTTFVVIANLLGIAVSFLVLTVLFPSPSVF
MAB_1459c|M.abscessus_ATCC_199 WLLGTADESTFIQRIRIQLILTVFVVAANLIGIGVVILLVTVAFPTPNVF
MLBr_00201|M.leprae_Br4923 WGIRTPWPLFSLSMLQSDIIGALFVLGFLRYGLPPQDRVELQDLPIRNLA
: :: : : : :: .: : :* .:
Rv1318c|M.tuberculosis_H37Rv RDTPRWLTFGVVPGYVLLALALGSYALTRQTVQALRWAIEGRKPTREEER
Mb1352c|M.bovis_AF2122/97 RDTPRWLTFGVVPGYVLLALALGSYALTRQTVQALRWAIEGRKPTREEER
MMAR_4078|M.marinum_M TEAPWWITFVVVPSYIVVALALGTYWITRRTVLALRWAIEERQPTPEDER
MAV_1540|M.avium_104 NDAPAWITFGVGPAYVASALAVGTYWITRRTVLALRWAIEERKPTPTDER
MUL_3944|M.ulcerans_Agy99 TEAPRWLTFGVGAGYDVTAFVVGSFWITSNTVRALRWYIEGRPPNRAVSR
Mflv_2336|M.gilvum_PYR-GCK SDAPAWLTFGVTPAYMLVALFFGTAWITSRTVRSLRWAIEERQPTSTDQR
Mvan_4309|M.vanbaalenii_PYR-1 SDAPAWLTFGVVPAYMVVALVLGTAWITSRTVGSLRWAIEERSPTRTDQR
TH_3474|M.thermoresistible__bu SDAPAWLTFGVSPALVGVALVVGTTWITRGIVRNLRWAIEERPPSRTDQR
MSMEG_4924|M.smegmatis_MC2_155 DARVAPYTYIVAPAYVVGALVVGVIWATNRVIGNVRWALEERPPTPQDQR
MAB_1459c|M.abscessus_ATCC_199 -DAPAWVNFVIVPVYIALAVLIGAVWGTRRIFKAVRWALDERPPTKADQR
MLBr_00201|M.leprae_Br4923 ISTLSVVVAFSTGFAVNLKLLMPVFRWQR---RDNRLAEADPAATELARS
. . * ..
Rv1318c|M.tuberculosis_H37Rv RTFLAPWRVAVGHLMFWGVGTALLTTLYGLINNAFIPRFLFAVSFCGVLV
Mb1352c|M.bovis_AF2122/97 RTFLAPWRVAVGHLMFWGVGTALLTTLYGLINNAFIPRFLFAVSFCGVLV
MMAR_4078|M.marinum_M NTFLAAWRVAVVDLILWGSGAVLLTTLYGMANTMFIPRFLFAVTFCGVLV
MAV_1540|M.avium_104 NTFLAPWRIALVDLVLWGAGTLLLTVLYGLVDTMFIPRFLFGVSFCGVLV
MUL_3944|M.ulcerans_Agy99 ETFLVPSRIAAVHLFLWGLGTLLFTTLYGLIDIRFIPKYLFSVSFCGILV
Mflv_2336|M.gilvum_PYR-GCK NVFLAPARVAQKLLLLWGVGAVLLTVLYGLQDTAFIPRFLFAVGFPGIVV
Mvan_4309|M.vanbaalenii_PYR-1 NVFVAPSRVAQMLLLFWGFGAALLTLLYGLQDSAFIPRFLFAVGFPGIVV
TH_3474|M.thermoresistible__bu NTFLAPWRVARAHLVLWGAGTAVLTTLYGLVDPAFIPRYLFTVSFAGIVV
MSMEG_4924|M.smegmatis_MC2_155 NTFFAPFRLTRVLLILWAAGTALLTTLYGMVDTNFIPKVLLGISFPGIVV
MAB_1459c|M.abscessus_ATCC_199 NAFAVPWRLTVIQSVLWAGGVALFTLLYGLYDPMYIPKIFFAVGFSGAVV
MLBr_00201|M.leprae_Br4923 RALRMPFYHTIISLVTWGIGGAVFIIASWSAARHSAPIVALATALGATAT
..: . : . *. * :: * : : . .
Rv1318c|M.tuberculosis_H37Rv ATATYLHTEFALRPFAAQALEAGPPPRRLAPGILGRTMVVWLLGSGVPVV
Mb1352c|M.bovis_AF2122/97 ATATYLHTEFALRPFAAQALEAGPPPRRLAPGILGRTMVVWLLGSGVPVV
MMAR_4078|M.marinum_M ATGSYLLTEFALRPVAAQALAAGPPPRRLSSGIMGRTMMVWLLGSGVPVV
MAV_1540|M.avium_104 ATACYLLAEFALRPVAAQALEAGPPPRRLTAGIMGRTMTVWFLGSGVPVI
MUL_3944|M.ulcerans_Agy99 ATGSYLLTEFALRPLTAQALETGPPPRRIVSGIMGRTILIWLLGSGVPVV
Mflv_2336|M.gilvum_PYR-GCK ATACYMITEFALRPIAAQALEAGRPPRRLAHGVMGRTMTVWLLSSGVPVL
Mvan_4309|M.vanbaalenii_PYR-1 ATACYMITEFALRPVAAQALEAGRPPRRLAHGVMGRTMTVWLLSSGVPVL
TH_3474|M.thermoresistible__bu ATICYLFTEFALRPVAAQALEAGPPPRRLAHGIMGRTMTVWLLGSGVPLL
MSMEG_4924|M.smegmatis_MC2_155 SVSCHLFTEFALRPVAAQALEAGPPPVRLAPGIMGRTMVVWALGSGVPIL
MAB_1459c|M.abscessus_ATCC_199 CAANYLFTEFAMRPVAARALEAGYRRRRLASGVMVRTVLAWMLGSGVPAA
MLBr_00201|M.leprae_Br4923 AIIGYLQSERVLRPVAVAALRSGVPENVKTPGVTLRLMLTWVPSTGVPVL
. :: :* .:**.:. ** :* *: * : * .:***
Rv1318c|M.tuberculosis_H37Rv GIALMAMFE-MVLLNLTRMQFATGVLIISMVTLVFGFILMWILAWLTATP
Mb1352c|M.bovis_AF2122/97 GIALMAMFE-MVLLNLTRMQFATGVLIISMVTLVFGFILMWILAWLTATP
MMAR_4078|M.marinum_M GIALVAIFQ-IALRNLSETQFAVGVLIVSMATLIFGFILMWIMSWLTATP
MAV_1540|M.avium_104 GIALLALFE-IWLRNLTETEFAVGVLIVSTAALIFGCLLMWILAWLTATP
MUL_3944|M.ulcerans_Agy99 DIALLAVFE-IALLHLSEVEFAVGVLIASSATLVFGLLLIAISAWLTATP
Mflv_2336|M.gilvum_PYR-GCK GILLLAVFS-LSLKNLSATQFAVAVIFIAAFALVFGLTLMWLLSWLIATP
Mvan_4309|M.vanbaalenii_PYR-1 GILLLAIFT-LSLRNLSATQFAVAVMIISVVALMFGLILMWLLSWLIATP
TH_3474|M.thermoresistible__bu GIMIAAAVS-LTQRHLSLVQFTIATMVLAAVALVFGFVLMWLVSWLTATP
MSMEG_4924|M.smegmatis_MC2_155 GILLAGVFA-VTLENLTPTQFAYAVMGLAVFALVFGFILMWILSWLTATP
MAB_1459c|M.abscessus_ATCC_199 GIVITGIGA-LVLHNLTVSQLAVAVIIIAGAIPSFGLILIWIASWLMATP
MLBr_00201|M.leprae_Br4923 AIVLAVVADKIALLHAVPEQLFSPILLLALAVLVVGLVSTWLVAMSIADP
* : : : :: : : .* : : * *
Rv1318c|M.tuberculosis_H37Rv VRVVRAALRRVERGELRTNLVVFDGTELGELQRGFNAMVAGLRERERVRD
Mb1352c|M.bovis_AF2122/97 VRVVRAALRRVERGELRTNLVVFDGTELGELQRGFNAMVAGLRERERVRD
MMAR_4078|M.marinum_M VRVVRAALNRVANGDLRGDLVVFDGTELGELQRGFNTMVDGLRERERVRD
MAV_1540|M.avium_104 VRVVRAALKRVERGDLRGDLVVFDGTELGELQRGFNAMVDGLRERERVRD
MUL_3944|M.ulcerans_Agy99 VRVVRAALKRVERGDLRGDVVVFDGSELGELQSGFNSMVEGLRERERVRD
Mflv_2336|M.gilvum_PYR-GCK VRVVRSALKQVEDGDLDCNLVVFDGTELGELQRGFNSMVHGLRERERVRD
Mvan_4309|M.vanbaalenii_PYR-1 VRVVRSALKHVEDGDLDCNVVVFDGTELGELQRGFNSMVHGLRERERVRD
TH_3474|M.thermoresistible__bu VRVVREALKQVENGNLDCNVVVFDGTELGELQRGFNAMVDGLRERERVRD
MSMEG_4924|M.smegmatis_MC2_155 IRVVRAALNRVEQGDLDTNLVVFDGTELGQLQRGFNSMVHGLRERERVRD
MAB_1459c|M.abscessus_ATCC_199 VKEVQTAIKRVAQNDLDIDLVVYDGTELGELQAGFNTMVEGLRERERVRD
MLBr_00201|M.leprae_Br4923 LRQLRWALSEVQRGNYNAHMQIYDASELGLLQAGFNDMVRELSERQRLRD
:: :: *: .* .: .: ::*.:*** ** *** ** * **:*:**
Rv1318c|M.tuberculosis_H37Rv LFGRHVGREVAAAAERERSKLGGEERHVAVVFIDIVGSTQLVTSRPPADV
Mb1352c|M.bovis_AF2122/97 LFGRHVGREVAAAAERERSKLGGEERHVAVVFIDIVGSTQLVTSRPPADV
MMAR_4078|M.marinum_M LFGRHVGREVAIAAERERPKLGGEERHVAVVFIDIVGSTKLVTSQPPADV
MAV_1540|M.avium_104 LFGRHVGREVALAAERERPKLGGEERHVAVVFIDIVGSTQLVTRRPPAEV
MUL_3944|M.ulcerans_Agy99 LFGRHVGREVAAAAERKPPQLGGEERYVAVVFVDIIGSTQLVSNRPATDV
Mflv_2336|M.gilvum_PYR-GCK LFGRHVGREVALAAEKQQVELGGEERRAAVIFVDIIGSTKLVTSRPAAEV
Mvan_4309|M.vanbaalenii_PYR-1 LFGRHVGREVARAAEKQKIELGGEERHAAVIFVDIIGSTQLVTSRPAVEV
TH_3474|M.thermoresistible__bu LFGRHVGREVARAAEEQQLELGGEERHVAVIFVDIVGSTQLVTTRPAAEV
MSMEG_4924|M.smegmatis_MC2_155 LFGRHVGREVAVLAEQQKFTLGGEERHAAVVFVDVIGSTQLVTSRPAVEV
MAB_1459c|M.abscessus_ATCC_199 LFGKHVGREVAAAAEQQQPELGGEERHIAVLFADIVGSTRLAATRPAVEV
MLBr_00201|M.leprae_Br4923 LFGRYVGEDVARRALEHGTELGGQERDVAVLFVDLIGSTQLAETKPPTKV
***::**.:** * .. ***:** **:* *::***:*. :*...*
Rv1318c|M.tuberculosis_H37Rv VKLLNKFFAIVVDEVDRHHGLVNKFEGDASLTIFGAPNRLPCPEDKALAA
Mb1352c|M.bovis_AF2122/97 VKLLNKFFAIVVDEVDRHHGLVNKFEGDASLTIFGAPNRLPCPEDKALAA
MMAR_4078|M.marinum_M VALLNRFFAVVVDEVDRHRGLVNKFEGDASLAIFGAPNHLERPEDQALAA
MAV_1540|M.avium_104 VSVLNQFFGIVVEEVDRHCGLVNKFEGDATLAIFGAPNHLDCPEDAALTA
MUL_3944|M.ulcerans_Agy99 VQLLDRFFAVIVDEVERHRGLVNKFEGDASLAIFGAPNPLECPEVAALSA
Mflv_2336|M.gilvum_PYR-GCK VELLNRFFDVVVDEVDKHHGLVNKFEGDAVLAVFGAPVSLDNAEAEALAA
Mvan_4309|M.vanbaalenii_PYR-1 VELLNRFFDVVVEEVDNHHGLVNKFEGDAVLAVFGAPVSLDSAETEALSA
TH_3474|M.thermoresistible__bu VSLLNRFFAVIVEEVDRHRGLVNKFEGDATLAIFGAPNRLDNPEDAALAA
MSMEG_4924|M.smegmatis_MC2_155 VELLNRFFAVIVDEVDAHRGLVNKFEGDAVLAVFGAPVALDNAEDEALGA
MAB_1459c|M.abscessus_ATCC_199 VALLNRFFTVVVEEIDRYEGMVNKFEGDATLAVFGAPVRQDRPESQALAA
MLBr_00201|M.leprae_Br4923 VHLLNEFFRVVVDTVGRHGGFVNKFQGDAALAIFGAPIEHPDSAGAALSA
* :*:.** ::*: : : *:****:*** *::**** . ** *
Rv1318c|M.tuberculosis_H37Rv ARAIADRLVNEMPECQAGIGVAAGQVIAGNVGARERFEYTVIGEPVNEAA
Mb1352c|M.bovis_AF2122/97 ARAIADRLVNEMPECQAGIGVAAGQVIAGNVGARERFEYTVIGEPVNEAA
MMAR_4078|M.marinum_M ARTIAARLASEVPECQAGIGVAAGQVVAGNVGSKERFEYTVIGEPVNEAA
MAV_1540|M.avium_104 ARAIADRLANEMPQCRAGIGVAAGQVVAGNVGARERFEYTVIGEPVNEAA
MUL_3944|M.ulcerans_Agy99 ARAIAGRLCTEVPECPAGIGVAAGQVVAGNVGANERFEYTVIGGPVNEAA
Mflv_2336|M.gilvum_PYR-GCK ARAMSRRLAAEVPECTAGIGVASGQVVAGNVGAKERFEYTVIGEPVNEAA
Mvan_4309|M.vanbaalenii_PYR-1 ARAMARRLRTEVPECPAGIGVAAGQVVAGNVGAKQRFEYTVIGEPVNEAA
TH_3474|M.thermoresistible__bu ARAIAARLRTEVPEVRAGIGVASGQVVAGNVGARERFEYTVIGEPVNEGA
MSMEG_4924|M.smegmatis_MC2_155 ARGIARRLREEVPELDAGIGVAAGQVVAGNVGAKQRFEYTVIGEPVNEAA
MAB_1459c|M.abscessus_ATCC_199 ARAIARRLRTEVPECQAGIGVASGQAVAGNIGAHDRFEYTVIGDPVNEAA
MLBr_00201|M.leprae_Br4923 ARELHDELLPVLGSAEFGIGVSAGRAIAGHIGAQARFEYTVIGDPVNEAA
** : .* : . ****::*:.:**::*:. ******** ****.*
Rv1318c|M.tuberculosis_H37Rv RLCELAKSRPGKLLASAQAVDAASEEERARWSLGRHVKLRGHDQPVRLAK
Mb1352c|M.bovis_AF2122/97 RLCELAKSRPGKLLASAQAVDAASEEERARWSLGRHVKLRGHDQPVRLAK
MMAR_4078|M.marinum_M RLCELAKDHPGRLLASGAALDGASHDERARWSVGKTVTLRGHDQPTRLAA
MAV_1540|M.avium_104 RLCELAKSHSGRLLATGDAIEGASEKERARWSLGDTVTLRGHERPTRLAS
MUL_3944|M.ulcerans_Agy99 RLCELAKSYSTRLVASSETVAGATETERAHWNLAETVTLRGRDRPTRLAI
Mflv_2336|M.gilvum_PYR-GCK RLCELSKKIDGHLVASADAVHNATETERRHWELGETVTLRGHAEPTTLAT
Mvan_4309|M.vanbaalenii_PYR-1 RLCELAKNVPALLVASADAVGGATESERALWKLGETVTLRGHDQPTRLAT
TH_3474|M.thermoresistible__bu RLCELAKTKPGLLVASADAVANAGEQERAHWVLGETVTLRGHNQPTRLAL
MSMEG_4924|M.smegmatis_MC2_155 RLCEFAKSVPGHVVASSDTLANASESERARWSLGESVTLRGLDEPTRLAV
MAB_1459c|M.abscessus_ATCC_199 RLCELSKTVPGHLVASLDTVLRAHHREALHWRSGDTVTLRGRTEPTELAV
MLBr_00201|M.leprae_Br4923 RLTELAKLEDGHVLASAIAVSGALDAEALCWNVGEIVELRGRTAPTQLAR
** *::* ::*: :: * . * * . * *** *. **
Rv1318c|M.tuberculosis_H37Rv PVGLTKPRR-------
Mb1352c|M.bovis_AF2122/97 PVGLTKPRR-------
MMAR_4078|M.marinum_M PI--------------
MAV_1540|M.avium_104 PAGAADRPAGTD----
MUL_3944|M.ulcerans_Agy99 PASSPRP---------
Mflv_2336|M.gilvum_PYR-GCK PVDRD-----------
Mvan_4309|M.vanbaalenii_PYR-1 PAGEL-----------
TH_3474|M.thermoresistible__bu PA--------------
MSMEG_4924|M.smegmatis_MC2_155 PV--------------
MAB_1459c|M.abscessus_ATCC_199 PV--------------
MLBr_00201|M.leprae_Br4923 PLNLAVPEQITSEVTG
*