For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRYPLAVAQLGFQRARTEENKRQRAAALVEAARSLALETGVASVTLTAVAGRAGIHYSAVRRYFTSHKEV LLHLAAEGWARWSGTVCEQLGEPGPMSAPRVAEALANGLAADPLFCDLLANLHLHLEQEVDVDRVIEVKR TSIAAVIALVDAIESALPALGRSGAFDILLAAYSLAATLWQIANPPERLTDAYAEEPELLPPEWNLDFAA ALTRLLTATLLGLLAGSPCECRSPTR
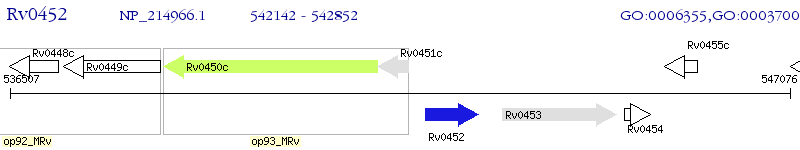
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0452 | - | - | 100% (236) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. tuberculosis H37Rv | Rv1534 | - | 6e-05 | 27.68% (177) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0460 | - | 1e-131 | 100.00% (236) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_3251 | - | 7e-06 | 30.43% (92) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2299c | - | 2e-83 | 69.72% (218) | transcriptional regulator |
| M. marinum M | MMAR_0773 | - | 1e-107 | 85.46% (227) | putative regulatory protein |
| M. avium 104 | MAV_4698 | - | 8e-87 | 77.89% (199) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2898 | - | 7e-05 | 35.71% (56) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3981 | - | 2e-06 | 28.66% (164) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_2569 | - | 7e-05 | 25.79% (159) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0452|M.tuberculosis_H37Rv -----------------MRYPLAVAQLGFQRARTEENKRQRAAALVEAAR
Mb0460|M.bovis_AF2122/97 -----------------MRYPLAVAQLGFQRARTEENKRQRAAALVEAAR
MMAR_0773|M.marinum_M MRKGTVPGRHYPRKSRSVRYPLAVAQLTFRRARTEENKRQRAAALVEAAR
MAV_4698|M.avium_104 ---------------------------------------------MEAAR
MAB_2299c|M.abscessus_ATCC_199 -----------------------MAQLTFQRARTDEKKRQRAAAIVEAAR
MUL_3981|M.ulcerans_Agy99 -----------------------MGGRRVKPGPRDE-KGVLAAQILSVAR
Mvan_2569|M.vanbaalenii_PYR-1 -----------------------MS--KIPPGPRDE-RGVLSSRILVAAR
MSMEG_2898|M.smegmatis_MC2_155 -----------------------------MPRVSREQMQSNRAAVVQAAS
Mflv_3251|M.gilvum_PYR-GCK -------------------MSAETRSSGRRKLPKQERSREMVAKIVQTTA
: .:
Rv0452|M.tuberculosis_H37Rv SLALETGVASVTLTAVAGRAGIHYSAVRRYFTSHKEVLLHLAAEGWARWS
Mb0460|M.bovis_AF2122/97 SLALETGVASVTLTAVAGRAGIHYSAVRRYFTSHKEVLLHLAAEGWARWS
MMAR_0773|M.marinum_M SMASETGVASVTLTAVASRAGIHYSAVRRYFTSHKEVLLHLAAEGWQRWS
MAV_4698|M.avium_104 SLALETGVASVTLTAVARRAGIHYSAVRRYFTSHKEVLLHLSAEGWVRWS
MAB_2299c|M.abscessus_ATCC_199 SLAVESGVTSVTLTAVANRAGVHYSAVRRYFSSHKEVLLHLAAEGWTRWS
MUL_3981|M.ulcerans_Agy99 DSFAENGSAGTTVRSVARAADVDPSLIYHYFGS-KEGLLDAATMPPQRWL
Mvan_2569|M.vanbaalenii_PYR-1 GAFADTGYAGTTIRAVARAADVDPALVYHYFGS-KEALLDAATDPPQRWL
MSMEG_2898|M.smegmatis_MC2_155 ELIRERGMADVTVDAISARAGLTHGSFHKQFASKQALLLEAICAAVDQRR
Mflv_3251|M.gilvum_PYR-GCK ALLIKTEPEQVTTNLIADRAGISKGSIYQYFSDKEEIIEAAVEHLAAQQA
. .* :: *.: . . : * . : : :
Rv0452|M.tuberculosis_H37Rv GTVCEQLGEPGPMSAPRVAEALANGLAADPLFCDLLANLHLHLEQEVDVD
Mb0460|M.bovis_AF2122/97 GTVCEQLGEPGPMSAPRVAEALANGLAADPLFCDLLANLHLHLEQEVDVD
MMAR_0773|M.marinum_M NTVCGELSQPGPMTESRVAAALADGLAADPLFCDLLANLHLHLEHEVEVD
MAV_4698|M.avium_104 NTVSDKLAQPGAATPSRIAETLADALADDPLFCDLLANLHLHLEHEVDAE
MAB_2299c|M.abscessus_ATCC_199 ATVCAALAEPGPGSPARVAESLVHGLVDDPLFCDLLANQHLHLEHEVDVE
MUL_3981|M.ulcerans_Agy99 DTVAETWTTPKADLGRRLLQTLLDSWADDEIGPSLRAILLTAAHEDATRL
Mvan_2569|M.vanbaalenii_PYR-1 QNVARAWTTPVPQLGAALVRLMLDAWADEEIGPVLRAILQTAAHEAATRE
MSMEG_2898|M.smegmatis_MC2_155 AAVQKTVEEHGSDAPDVLKDWYLSEEHRDNPATGCAVAALVGHAATVTDD
Mflv_3251|M.gilvum_PYR-GCK PEIEEMLRSVTMSRPEVAMESSIDILIDFTIANRRLIRYLAERPDHVRTF
: .
Rv0452|M.tuberculosis_H37Rv RVIEVKRTSIAAVIALVD-AIESALPALGRSGAFDILLAAYSLAATLWQI
Mb0460|M.bovis_AF2122/97 RVIEVKRTSIAAVIALVD-AIESALPALGRSGAFDILLAAYSLAATLWQI
MMAR_0773|M.marinum_M RVVEVKRTSTAAVIALAD-AIENALPALGRAGAFDVLLAAYSLGATLWQI
MAV_4698|M.avium_104 RVVEVKRISTAATLSLAD-SIQRALPELGRSGSLDILLAAYSLAATLWQV
MAB_2299c|M.abscessus_ATCC_199 RVAEVKQISNVAVMSIAE-SIERTLPEIGRKGSLDILVAGFSLAAMLWQI
MUL_3981|M.ulcerans_Agy99 KLRRIVESSLIGVSQLDT-DED---DRLTRSGLIASQLIGLAMTRYVWQI
Mvan_2569|M.vanbaalenii_PYR-1 KLRLVVERSLMGVSQLGV-DER---DRMLRSGLISSQIMGLAMMRYVWRI
MSMEG_2898|M.smegmatis_MC2_155 ALRDAYCRAVGDLLAMIE-DGG----TADAFGEVALMVGALELAR-----
Mflv_3251|M.gilvum_PYR-GCK EAISGLNATLLAMATLHMGHYRDQYRDELSPRALAWLFFNMAVATTLRYI
: : . . :
Rv0452|M.tuberculosis_H37Rv ANPPERLTDAYAEEPELLPPEWNLDFAAALTRLLTATLLGLLAGSPCECR
Mb0460|M.bovis_AF2122/97 ANPPERLTDAYAEEPELLPPEWNLDFAAALTRLLTATLLGLLAGSPCECR
MMAR_0773|M.marinum_M ANPPQRLTDVYAEEPDVLPPEWNLDFAAALTRLLTATCCGLTAKSS----
MAV_4698|M.avium_104 ANPPERLTDAYAEEPEVVPPEWNLDFASALTRLLTATCIGLIPQSA----
MAB_2299c|M.abscessus_ATCC_199 AHPPEGLEHDYRSEP-GVPPDWSLDFEPALTRLLTATCIGIVEQKH----
MUL_3981|M.ulcerans_Agy99 E-PIASMSDS--EVLAAITPNLQRYIDGDLHPTKNSPAQRPRRTRRP---
Mvan_2569|M.vanbaalenii_PYR-1 E-PVASMDDD--ELVAAVAPNLQRYISDDLG-------------------
MSMEG_2898|M.smegmatis_MC2_155 --------------ATKGTPLSARFLESARQRLSRQE-------------
Mflv_3251|M.gilvum_PYR-GCK EADDP-------ISLDELRSGLKFASTGLLAGAVSGGHVGEPGPS-----
.
Rv0452|M.tuberculosis_H37Rv SPTR
Mb0460|M.bovis_AF2122/97 SPTR
MMAR_0773|M.marinum_M ----
MAV_4698|M.avium_104 ----
MAB_2299c|M.abscessus_ATCC_199 ----
MUL_3981|M.ulcerans_Agy99 ----
Mvan_2569|M.vanbaalenii_PYR-1 ----
MSMEG_2898|M.smegmatis_MC2_155 ----
Mflv_3251|M.gilvum_PYR-GCK ----