For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PVLRRLGCSVLALGLLAGCAPPRTGPASSPTNNGAKADAVIRIVRDFMTQAHLKAVLVRVTVAGKEVVTR AVGDSMTGVPATTAMHFRNGAVAISYVATLLLKLVDEKKLRLDDKLSRWLPDFPHADRVTLGQLAQMTSG YPDYVLGNEAFDAELYANPFRQWTTQELLDQISSRPLLYDPGTNWNYAHTNYLLLGLALEKAAGQDMPTL LQRKVLSPLGLTATANSDTPAIPEPALHAFTSERRAALKIPAGVPFYEESTFWNPSWTITHGAIQTTTIY DMEATAVGIGSGRLLSADSYKKMVSTELRGKTRAQPGCPTCFEQNDGYSYGLGIVISGHWLLQNPMFAGY AAVEAYLPSQRVAVAVAVTYAPEAFDDQGNYRNQADILFRKIGAEVAPNDAPPMPPGR*
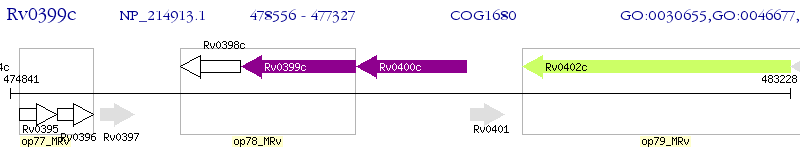
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0399c | lpqK | - | 100% (409) | POSSIBLE CONSERVED LIPOPROTEIN LPQK |
| M. tuberculosis H37Rv | Rv1922 | - | 1e-25 | 27.79% (385) | lipoprotein |
| M. tuberculosis H37Rv | Rv1730c | - | 3e-11 | 26.09% (299) | penicillin-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0405c | lpqK | 0.0 | 100.00% (409) | lipoprotein LpqK |
| M. gilvum PYR-GCK | Mflv_0495 | - | 5e-12 | 26.34% (224) | beta-lactamase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0330 | - | 1e-41 | 31.15% (366) | Beta-lactamase |
| M. marinum M | MMAR_0696 | lpqK | 0.0 | 79.18% (413) | lipoprotein, LpqK |
| M. avium 104 | MAV_3991 | - | 8e-11 | 26.45% (276) | beta-lactamase |
| M. smegmatis MC2 155 | MSMEG_1586 | - | 7e-37 | 32.66% (297) | beta-lactamase |
| M. thermoresistible (build 8) | TH_3520 | - | 2e-40 | 30.05% (366) | PUTATIVE beta-lactamase |
| M. ulcerans Agy99 | MUL_1772 | - | 3e-07 | 21.52% (330) | beta-lactamase |
| M. vanbaalenii PYR-1 | Mvan_3510 | - | 1e-167 | 70.49% (410) | beta-lactamase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0399c|M.tuberculosis_H37Rv -------------------MPVLRRLGCSVLALGLLAGCA---PPRTGPA
Mb0405c|M.bovis_AF2122/97 -------------------MPVLRRLGCSVLALGLLAGCA---PPRTGPA
MMAR_0696|M.marinum_M -------------------MPILRRVAGVVFGVCLLAGCAALHPPQQSSA
Mvan_3510|M.vanbaalenii_PYR-1 -------------------MTVLRRLAAACILP-ILAACA---PPAVAPD
MAB_0330|M.abscessus_ATCC_1997 ---------MPELSTRCFDESSLMRRILAVVLTIALLVSACGKHSAIVVA
MSMEG_1586|M.smegmatis_MC2_155 ------------MMSAWPLVPGFVRAATGAVIAAITFVAAS----ACATA
TH_3520|M.thermoresistible__bu ---------------------MRAVRTLAALLLAVFAVSAG-----PSAA
Mflv_0495|M.gilvum_PYR-GCK MTRSSTRLRRLGAAGAVVLALGACGDRPEPPGTSAVRPPATPTRVLPSPP
MAV_3991|M.avium_104 -----------------MLTLVAASSTGRVVHTS---PSEAPPASMTPHA
MUL_1772|M.ulcerans_Agy99 ----------------MGVARPRWRGPRAAWFAGARKPSDG-----RRLT
Rv0399c|M.tuberculosis_H37Rv SS-PTNNGAKAD--AVIRIVRDFMTQAHLKAVLVRVTVAGKEVVTRAVG-
Mb0405c|M.bovis_AF2122/97 SS-PTNNGAKAD--AVIRIVRDFMTQAHLKAVLVRVTVAGKEVVTRAVG-
MMAR_0696|M.marinum_M PTGPDGKGTKAD--AVMKVVRDFMGPAHLRSVIVRVSVDGKEIVTDAVG-
Mvan_3510|M.vanbaalenii_PYR-1 SDAPKADPAKAD--AVMRIVRETMAEAHLKAVIVRVTVDGREVLTEAAG-
MAB_0330|M.abscessus_ATCC_1997 ELDPQTTAKLDS--AINEILTSAAVPGAIVGVWGP---KGQYVRTFGVA-
MSMEG_1586|M.smegmatis_MC2_155 PAAPPIAAQLDD--AVGKVMADLSIPGAVVGFWGP---DVTYVRAFGVA-
TH_3520|M.thermoresistible__bu GQPPDLAAALDR--AIELRLDQMGVPGAIVSLSIPG--KIEYTRAFGVG-
Mflv_0495|M.gilvum_PYR-GCK PPVPEVT-PTGDFSAVARLVEDAIAARRLPGAVVQVGHAGKVVFRRAFGV
MAV_3991|M.avium_104 SLVPESTSPAEDFTTISKLVNDAIAARGLPGAVVMVGHGGKVVFHQAYGS
MUL_1772|M.ulcerans_Agy99 RRPPAANLNPVNLDANQADIREVCDAGLLSGAVTVVWQHGEVLQVNEIG-
* : : . : . .
Rv0399c|M.tuberculosis_H37Rv -----------DSMTGVPATTAMHFRNGAVAISYV-ATLLLKLVDEKKLR
Mb0405c|M.bovis_AF2122/97 -----------DSMTGVPATTAMHFRNGAVAISYV-ATLLLKLVDEKKLR
MMAR_0696|M.marinum_M -----------DSMTGVPASTDMHFRNGAVAISYV-ATLLLKLVDEHKLS
Mvan_3510|M.vanbaalenii_PYR-1 -----------ESMTGVPANTAMHFRNGAVAISYV-ATLLLKLAEEGTVS
MAB_0330|M.abscessus_ATCC_1997 -----------NTATRAPLRPDFYHRIGSVTKTFT-VTAVLQLVDEGKVE
MSMEG_1586|M.smegmatis_MC2_155 -----------DTATRAPMQADMYTRIGSLTKTFT-ITALLTLIDQGKAG
TH_3520|M.thermoresistible__bu -----------DTATGTPMLVDNHHRIGSVTKTFI-GTAILQLADQGRIR
Mflv_0495|M.gilvum_PYR-GCK RKLESEPGLDGAPAPAEPMTEDTVFDIASLTKSLATATAVMQLHEQGRVR
MAV_3991|M.avium_104 RKLAGEPGLDGAPAPAEPMTEDTIFDLASLTKCLATAVAVMQLYEQGKVA
MUL_1772|M.ulcerans_Agy99 ---------YRDVEAGLPMQRDTLFRIASMTKPVT-VAAAMSMVDEGKIA
. * .::: . : : ::
Rv0399c|M.tuberculosis_H37Rv LDDKLSRWLPDF-----------------PHADRVTLGQLAQMTSGYPDY
Mb0405c|M.bovis_AF2122/97 LDDKLSRWLPDF-----------------PHADRVTLGQLAQMTSGYPDY
MMAR_0696|M.marinum_M LDDKLSKWLPDF-----------------PHADRVTLGQLAQMTSGYPDY
Mvan_3510|M.vanbaalenii_PYR-1 LDDKLSTWLPRI-----------------PHADRVTLRQLAQMTSGYVDY
MAB_0330|M.abscessus_ATCC_1997 LDDPIAKYVDKV-----------------PNGNKITLRELARMQSGLFNY
MSMEG_1586|M.smegmatis_MC2_155 LDDPISMYVADV-----------------PSGDTITLRHLAQMQSGLVTY
TH_3520|M.thermoresistible__bu LSDPISRYVEGV-----------------PSGDVITLDMLARMRSGLADY
Mflv_0495|M.gilvum_PYR-GCK IDEPVQTYLPGFNPA------------GDPRRAAVTVRMLLTHTSGIAGD
MAV_3991|M.avium_104 FDDPVQKYLPAFNTT------------NDPQRAKVTIRMLLTNTSGEGID
MUL_1772|M.ulcerans_Agy99 LRDPITRWAPELRDIRVLDDPHGPLDRTHPTRRPILIEDLLTHTSGLAYS
: : : : . * : : * **
Rv0399c|M.tuberculosis_H37Rv VLGNEAFDAELYANPFR-----QWTTQELLDQISSRPLLYDPGTNWNYAH
Mb0405c|M.bovis_AF2122/97 VLGNEAFDAELYANPFR-----QWTTQELLDQISSRPLLYDPGTNWNYAH
MMAR_0696|M.marinum_M VLGNDEFGDALYADPFR-----QWTPQELLAQISSRPLLYEPGTNWNYAH
Mvan_3510|M.vanbaalenii_PYR-1 VIGTTAMNDAFLADPFR-----AWTTHDLLSYATSKPLLYEPGTNWNYAH
MAB_0330|M.abscessus_ATCC_1997 SM-SLAFSRDLEADPRR-----QYSTHELLDYAFAQPSNFVPGQGYEYSN
MSMEG_1586|M.smegmatis_MC2_155 DG-VPEFEAAFLADPQR-----TFTPQQLLGYALDKPLQFPPGTQYDYCN
TH_3520|M.thermoresistible__bu TE-TDEFVERLYRESPSGPDAFAVTPRELVESAFARPLNFAPGSEFEYSN
Mflv_0495|M.gilvum_PYR-GCK LSLDGPWG---LAGPDK---------ADGVRRALGAWVVSDPGEGFHYSD
MAV_3991|M.avium_104 VSLRDPWG---LAGPDK---------AEGIHRALTTPLQSAPAGEFRYSD
MUL_1772|M.ulcerans_Agy99 FSVSGDISRAYMRLPFG------HGSDAWLAELAALPLVHQPGERVTYSH
. : *. *..
Rv0399c|M.tuberculosis_H37Rv TNYLLLGLALEKAAGQDMPTLLQRKVLSPLGLTATANSDTPAIPEP----
Mb0405c|M.bovis_AF2122/97 TNYLLLGLALEKAAGQDMPTLLQRKVLSPLGLTATANSDTPAIPEP----
MMAR_0696|M.marinum_M TNYVLLGLALEKATGQDMATLLKQKVLAPLGLTATANSETPSIPDP----
Mvan_3510|M.vanbaalenii_PYR-1 TNYVLLGLALEKATGETMSDLLSDKVLRPLGLTDTAASLTADIPSP----
MAB_0330|M.abscessus_ATCC_1997 TNAVLLGQVVEKVSGQTLPAFVQEHITGPLGMRRTSFPETSAIADP----
MSMEG_1586|M.smegmatis_MC2_155 TNTVLLGLVVEKLSGQSLADYVRQHILVPLKLTHTSIPTSPAFPEP----
TH_3520|M.thermoresistible__bu TNTVLLGMVVERVSGVPLARYLQDNIFGPVGLTQTSYPADGRLPAP----
Mflv_0495|M.gilvum_PYR-GCK INFIILGALVEKITGEPEDSYVDRNVFAPLKMSETRYLPVTKVCGPHRVR
MAV_3991|M.avium_104 INYLLLGALLEKITGEDEDDYVQHHVFGPLGMHDTRYLPPAKACGPHIIR
MUL_1772|M.ulcerans_Agy99 A-IDLLGVIMSRIDDKPFYQVLDERILGPAGMTDTGFFVSTQAQRR----
:** :.: . : .: * : *
Rv0399c|M.tuberculosis_H37Rv -----------------------------ALHAFTSERRAALKIPAGVPF
Mb0405c|M.bovis_AF2122/97 -----------------------------ALHAFTSERRAALKIPAGVPF
MMAR_0696|M.marinum_M -----------------------------ALHAFSAERRAFLKIPADTPF
Mvan_3510|M.vanbaalenii_PYR-1 -----------------------------VLHAFTSERREALRIPPDTPF
MAB_0330|M.abscessus_ATCC_1997 -----------------------------HARGYTQLTP--------TGS
MSMEG_1586|M.smegmatis_MC2_155 -----------------------------HPQGYTVLD----------GT
TH_3520|M.thermoresistible__bu -----------------------------YTHGYNQGP---------DGA
Mflv_0495|M.gilvum_PYR-GCK GTAIAADPHGQAVDNCPDGTWNIDLLERIAPTALDEDT---PGLNPDYGY
MAV_3991|M.avium_104 GAAIAWAPNGEPMTECPEWTWRTTLLPRIAPTARDEESRDDPSKNPDYYA
MUL_1772|M.ulcerans_Agy99 -----------------------------AATMYRLDELDQLR----HDV
Rv0399c|M.tuberculosis_H37Rv YEESTFWNPSWTITHGAI-----QTTTIYDMEATAVGIGSGRLLSADSYK
Mb0405c|M.bovis_AF2122/97 YEESTFWNPSWTITHGAI-----QTTTIYDMEATAVGIGSGRLLSADSYK
MMAR_0696|M.marinum_M SEESTFWNPSWTITHGAI-----QTTTIYDMEATAVGIGSGKLLSADSYQ
Mvan_3510|M.vanbaalenii_PYR-1 YEESTFWNPSWTITHGAI-----QTTNIYDLEATAVGIGSGRLLKPESYR
MAB_0330|M.abscessus_ATCC_1997 LVDSTDWNPSWASWAGAM-----IST-LDDLRIWVPALATGVLLKPDSQQ
MSMEG_1586|M.smegmatis_MC2_155 PRVTTDWNPSWAWSNGNM-----IST-LDDMRVWAQKLSSGELLSENLRD
TH_3520|M.thermoresistible__bu IVDTTLWNPAWADAAGKI-----VSN-VHDMRTWARALGSGALLRPDTHA
Mflv_0495|M.gilvum_PYR-GCK PLRGTVHDPTARRMGGVAG-SAGVFTTVHDMGRFAQALLDRVAGRPSPFP
MAV_3991|M.avium_104 LLRGTVHDPTARRMGGVAG-NAGVFSTAHDVGVYAQALLDRLAGRPSDFP
MUL_1772|M.ulcerans_Agy99 MGPPHVRPPSFCNAGGGLWSTADDYLRFVRLLLGDGTIDGVRVLSPESVR
*: * : : .
Rv0399c|M.tuberculosis_H37Rv KMVSTELRGKTRAQPG------------------------CPTCFEQNDG
Mb0405c|M.bovis_AF2122/97 KMVSTELRGKTRAQPG------------------------CPTCFEQNDG
MMAR_0696|M.marinum_M KMVSTALRGKTRAQPD------------------------CPTCFEQSEG
Mvan_3510|M.vanbaalenii_PYR-1 QMVSTDLRGKTRSQPG------------------------CATCRPMDER
MAB_0330|M.abscessus_ATCC_1997 QRLQTVA------------------------------------MPPVPDD
MSMEG_1586|M.smegmatis_MC2_155 ERFRT-A------------------------------------LP-MSEN
TH_3520|M.thermoresistible__bu ERIHG--------------------------------------GSSVVPG
Mflv_0495|M.gilvum_PYR-GCK LSQAGARMMTTPQQP-----------------AP------GRDLRGFGWD
MAV_3991|M.avium_104 LQQGTVELMTTPQQPGHDPRQLEAANRAARSGTPNYPAIKGQNLFGFGWD
MUL_1772|M.ulcerans_Agy99 LMRTDRLSDEHKRHNFLG--------------------APFWVGRGFGLN
Rv0399c|M.tuberculosis_H37Rv YSYGLGIVISGHWLLQN----PMFAGYAAVEAYLPS-QRVAVAVAVTYAP
Mb0405c|M.bovis_AF2122/97 YSYGLGIVISGHWLLQN----PMFAGYAAVEAYLPS-QRVAVAVAVTYAP
MMAR_0696|M.marinum_M YTYGLGIVISGHWLLQN----PLFSGYAATEAYLPS-QRVAIAVALTYTP
Mvan_3510|M.vanbaalenii_PYR-1 YTYGLGVVISGNWLLQN----PMFAGYAGVEAYLPA-QKIAIAVATTFAP
MAB_0330|M.abscessus_ATCC_1997 IRYGLGIFDARGWIGHS----GSIPGYQTLAIYSPAEQTTVVALLNTDVA
MSMEG_1586|M.smegmatis_MC2_155 AKYGVGAFDTNGWIGHS----GITPGFETVVVGLPEAQSTLVVFVNTDVP
TH_3520|M.thermoresistible__bu VDYSFAIFNAQGWLGHN----GDIPGYATVVVYLPERDATLVVFVNSDVP
Mflv_0495|M.gilvum_PYR-GCK IDTAHSGPRGSVFPVGS----FGHTGFTGVSVWLDPESDTYVVILGNVIH
MAV_3991|M.avium_104 IDTGYSRPRGRVFPIGS----FGNTGFTGTTLWMDPGSDTYVILLSNSIH
MUL_1772|M.ulcerans_Agy99 LSVVTDPVQSTPLFGPGGLGTFSWPGAYGTWWQADPGADLILLYLIQHCP
. .* :
Rv0399c|M.tuberculosis_H37Rv EAFDDQGNYRNQADILFRKIGAEVAPNDAPPMPPGR---
Mb0405c|M.bovis_AF2122/97 EAFDDQGNYRNQADILFRKIGAEVAPNDAPPMPPGR---
MMAR_0696|M.marinum_M EAFDDEGNYPNQADILFRKIAAALVPDDAPPMPPGR---
Mvan_3510|M.vanbaalenii_PYR-1 EAFDDSGDYRNEAETLFRRIGAEMAPDDAPPGPAPR---
MAB_0330|M.abscessus_ATCC_1997 KQPAQPSTLFGTAITKVISPDHVFLLENAIPQPR-----
MSMEG_1586|M.smegmatis_MC2_155 HEVG---TAVATAVTGVVTPDNVYR--------------
TH_3520|M.thermoresistible__bu EPHS--AGQIAYVVTEMVTPEHIYELGPQPPQMLDEPAA
Mflv_0495|M.gilvum_PYR-GCK QRGGPPITPLSGAVATEAARALHLYGS------------
MAV_3991|M.avium_104 LRGSPPISNLRGDVATAAARALHLYGAAPP---------
MUL_1772|M.ulcerans_Agy99 DLSVNAAAAVAGNPGLAKLRTAQPRFVRRTYRALGL---