For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRSSTRLRRLGAAGAVVLALGACGDRPEPPGTSAVRPPATPTRVLPSPPPPVPEVTPTGDFSAVARLVE DAIAARRLPGAVVQVGHAGKVVFRRAFGVRKLESEPGLDGAPAPAEPMTEDTVFDIASLTKSLATATAVM QLHEQGRVRIDEPVQTYLPGFNPAGDPRRAAVTVRMLLTHTSGIAGDLSLDGPWGLAGPDKADGVRRALG AWVVSDPGEGFHYSDINFIILGALVEKITGEPEDSYVDRNVFAPLKMSETRYLPVTKVCGPHRVRGTAIA ADPHGQAVDNCPDGTWNIDLLERIAPTALDEDTPGLNPDYGYPLRGTVHDPTARRMGGVAGSAGVFTTVH DMGRFAQALLDRVAGRPSPFPLSQAGARMMTTPQQPAPGRDLRGFGWDIDTAHSGPRGSVFPVGSFGHTG FTGVSVWLDPESDTYVVILGNVIHQRGGPPITPLSGAVATEAARALHLYGS
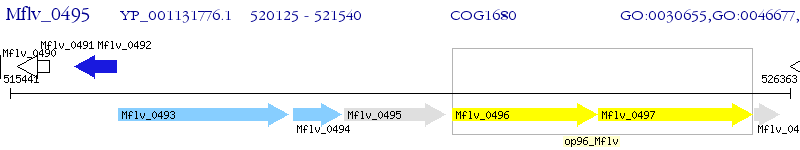
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0495 | - | - | 100% (471) | beta-lactamase |
| M. gilvum PYR-GCK | Mflv_3749 | - | 8e-19 | 27.70% (408) | beta-lactamase |
| M. gilvum PYR-GCK | Mflv_3473 | - | 4e-17 | 29.83% (238) | beta-lactamase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1759c | - | 1e-20 | 31.71% (205) | penicillin-binding protein |
| M. tuberculosis H37Rv | Rv1730c | - | 1e-20 | 31.71% (205) | penicillin-binding protein |
| M. leprae Br4923 | MLBr_00119 | lipE | 4e-07 | 34.23% (111) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_4800 | - | 1e-166 | 60.95% (484) | putative beta-lactamase |
| M. marinum M | MMAR_3399 | - | 1e-145 | 58.42% (457) | hypothetical protein MMAR_3399 |
| M. avium 104 | MAV_3991 | - | 1e-160 | 62.04% (461) | beta-lactamase |
| M. smegmatis MC2 155 | MSMEG_6805 | - | 0.0 | 70.44% (477) | beta-lactamase |
| M. thermoresistible (build 8) | TH_4016 | - | 1e-166 | 66.51% (436) | PUTATIVE beta-lactamase |
| M. ulcerans Agy99 | MUL_1772 | - | 1e-13 | 27.23% (235) | beta-lactamase |
| M. vanbaalenii PYR-1 | Mvan_5895 | - | 0.0 | 64.65% (495) | beta-lactamase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0495|M.gilvum_PYR-GCK ---------------------------------MTRS-STRLRRLGAAGA
MSMEG_6805|M.smegmatis_MC2_155 MSAGSAPLVTPRRKNALHAATASRTARYRRPVRLSRSRVPRLCAIGAVFA
MAB_4800|M.abscessus_ATCC_1997 -------------------------------MKVPGSHATRVCAVATVLL
Mvan_5895|M.vanbaalenii_PYR-1 ---------------------------------MPGSRVARLCAAGSLLL
TH_4016|M.thermoresistible__bu --------------------------------------------------
MMAR_3399|M.marinum_M --------------------------------------MPAMDAAG----
MAV_3991|M.avium_104 --------------------------------------MLTLVAASSTGR
Mb1759c|M.bovis_AF2122/97 --------------------------------MCPPIILSSATPTGTRCG
Rv1730c|M.tuberculosis_H37Rv --------------------------------MCPPIILSSATPTGTRCG
MUL_1772|M.ulcerans_Agy99 ----------------------------------MGVARPRWRGPRAAWF
MLBr_00119|M.leprae_Br4923 ------------------------------------MTSDGKIRVPTDLD
Mflv_0495|M.gilvum_PYR-GCK VVLALGACGDRPEPPGTSAVRPPATPTRVLPSPPPPVPE-VTPTGDFSAV
MSMEG_6805|M.smegmatis_MC2_155 VVAATSCSRPAPAPREPAPATGIAAASPAPTAPPTQVTQPVTPTGDFSAV
MAB_4800|M.abscessus_ATCC_1997 ALAGIPSCG-RPAQP--------------ITAPS------IVPTGPFAPI
Mvan_5895|M.vanbaalenii_PYR-1 TLATAPSCG-QPGHPTDPPPPSTGTPTATATATA------TDPVADFAAV
TH_4016|M.thermoresistible__bu -------------------------------------------------V
MMAR_3399|M.marinum_M -------------------------------------------LAAFAPV
MAV_3991|M.avium_104 VVHTSPSEA----------------PPASMTPHASLVPESTSPAEDFTTI
Mb1759c|M.bovis_AF2122/97 TRHGRAVVTEYVRALDR--------LPHEIATAVVETVNCADPGAAFDEL
Rv1730c|M.tuberculosis_H37Rv TRHGRAVVTEYVRALDR--------LPHEIATAVVETVNCADPGAAFDEL
MUL_1772|M.ulcerans_Agy99 AGARKPSDG---------------------RRLTRRPPAANLNPVNLDAN
MLBr_00119|M.leprae_Br4923 AVTAIGDED-----------------------------HSDIDSAAVERI
Mflv_0495|M.gilvum_PYR-GCK ARLVEDAIAARRLPGAVVQVGHAGKVVFRRAFGVRKLESEPGLDGAPAPA
MSMEG_6805|M.smegmatis_MC2_155 TRLVDDAVAARRLPGAVVQIGHAGKIVFRAAFGARKLDGEPGLDGSPSPA
MAB_4800|M.abscessus_ATCC_1997 TTLVNDAVAAPRLPGAVVEIGHAGKIVFRQAFGWRKLPDEPGLNGSPAPA
Mvan_5895|M.vanbaalenii_PYR-1 SQLMHDAIAAHRLPGAVVQIGHAGKIVFRQAFGSRKLAGEPGLDGSPAPA
TH_4016|M.thermoresistible__bu IGQVDDAVAVRRLPGAVVQIGHAGEVVFRQAFGSRKLAGEPGPTGEPTPA
MMAR_3399|M.marinum_M SRLINDAIAAERLPGAVVVVGHGGKIVFHQAYGSRKLAGEQGLDGVSAPA
MAV_3991|M.avium_104 SKLVNDAIAARGLPGAVVMVGHGGKVVFHQAYGSRKLAGEPGLDGAPAPA
Mb1759c|M.bovis_AF2122/97 DAKINAGMKAYAIPGVAVAV-WAGGQEYVKGYGVTNVDHPMPVDG-----
Rv1730c|M.tuberculosis_H37Rv DAKINAGMKAYAIPGVAVAV-WAGGQEYVKGYGVTNVDHPMPVDG-----
MUL_1772|M.ulcerans_Agy99 QADIREVCDAGLLSGAVTVVWQHGEVLQVNEIGYRDVEAGLPMQR-----
MLBr_00119|M.leprae_Br4923 WQATRHWYQAGMHPAIQLCIRQHGRVVLNRAIGHG--WGNAPTDPHDAEK
. .. : * *
Mflv_0495|M.gilvum_PYR-GCK EPMTEDTVFDIASLTKSLATATAVMQLHEQGRVRIDEPVQTYLPGFN---
MSMEG_6805|M.smegmatis_MC2_155 EPMTEDTLFDLASLTKSIATTTAVLQLYEQGKIRLDEPVQTYLPDFN---
MAB_4800|M.abscessus_ATCC_1997 EPMTDDTIFDIASLTKPLATSVAVLQLYEGGRVHIDEPVQAYLPDFN---
Mvan_5895|M.vanbaalenii_PYR-1 EPMTEDTIFDLASLSKSLATAPAFLQLYERGLVRIDEPVQTYLPEFN---
TH_4016|M.thermoresistible__bu EPMTEDTMFDLASLTKSLATTVAVLQLAEQGRLRVDDPVQSHLPEFN---
MMAR_3399|M.marinum_M EAMTEDTIFDIASLTKCLATATAIMQLYEQGKVGFDDPVQNHLPGFN---
MAV_3991|M.avium_104 EPMTEDTIFDLASLTKCLATAVAVMQLYEQGKVAFDDPVQKYLPAFN---
Mb1759c|M.bovis_AF2122/97 -----DTVFRIGSTTKTFTG-TVMMRLVERGKVDLDSPVRRYIPDFA---
Rv1730c|M.tuberculosis_H37Rv -----DTVFRIGSTTKTFTG-TVMMRLVERGKVDLDSPVRRYIPDFA---
MUL_1772|M.ulcerans_Agy99 -----DTLFRIASMTKPVTVAAAMSMVDE-GKIALRDPITRWAPELRDIR
MLBr_00119|M.leprae_Br4923 IPVTTDTPFCAYSAAKAITA-TVVHMLVERGHFSLDDRVCEYLPTYT---
** * * :* .: .. : * * . . . : *
Mflv_0495|M.gilvum_PYR-GCK ---------PAGDPRRAAVTVRMLLTHTSGIAGDLSLDGPWGLAGP----
MSMEG_6805|M.smegmatis_MC2_155 ---------PTGDPRRARVTLRMLLTHTSGIAGDLSLDGPWGLTAA----
MAB_4800|M.abscessus_ATCC_1997 ---------PTNDPRRDQVTLRMLLTHTSGIGGDLSHQGPWGLKQA----
Mvan_5895|M.vanbaalenii_PYR-1 ---------PAGDPRRAQVTLRMLLTHTSGLAGDLSLDGPWGLDKP----
TH_4016|M.thermoresistible__bu ---------PQHDPRRARVTLRMLLTHTSGIAGDLSLDGPWGLDRP----
MMAR_3399|M.marinum_M ---------PASDPQRAQVTVRMLLTQTSGEPVDVGLQDVWGLDGA----
MAV_3991|M.avium_104 ---------TTNDPQRAKVTIRMLLTNTSGEGIDVSLRDPWGLAGP----
Mb1759c|M.bovis_AF2122/97 ---------VADESASATVTVRQLLNHTAGWDGRNGQD--FGRGDD----
Rv1730c|M.tuberculosis_H37Rv ---------VADESASATVTVRQLLNHTAGWDGRNGQD--FGRGDD----
MUL_1772|M.ulcerans_Agy99 VLDDPHGPLDRTHPTRRPILIEDLLTHTSGLAYSFSVSGDISRAYMRLPF
MLBr_00119|M.leprae_Br4923 ------------SYGKHRTTIRHVLTHSAGVPFPTGPRPDVTRADD----
:. :*.:::* .
Mflv_0495|M.gilvum_PYR-GCK --DKADGVRRALGAWVVSDPGEGFHYSDINFIILG-ALVEKITGEPEDSY
MSMEG_6805|M.smegmatis_MC2_155 --DKAEGVKRALAAWVVFEPGAMFHYSDIGFIILG-TLVEKITGQSLDGY
MAB_4800|M.abscessus_ATCC_1997 --DKADGIHRALTTPLAFDPGTTFHYSDINFIILG-ALVEKVTGQTLDIY
Mvan_5895|M.vanbaalenii_PYR-1 --DKADGIRRALAAWVVFDPGEIFHYSDINFILVG-AIIEKLTGEQLDSY
TH_4016|M.thermoresistible__bu --DKAEGVRRALGAWVVHEPGRLFHYSDIGFILLG-AIVEKLTGEPLDVY
MMAR_3399|M.marinum_M --DRVEGIHRALSTPLQSPPGREFRYSDINYILLG-ALLEKIAGEAEDGY
MAV_3991|M.avium_104 --DKAEGIHRALTTPLQSAPAGEFRYSDINYLLLG-ALLEKITGEDEDDY
Mb1759c|M.bovis_AF2122/97 --AVALYVKAMTRLPQLTPPGTAFAYNNSGLVVAG-RIIELVAGTTYEST
Rv1730c|M.tuberculosis_H37Rv --AVALYVKAMTRLPQLTPPGTAFAYNNSGLVVAG-RIIELVAGTTYEST
MUL_1772|M.ulcerans_Agy99 GHGSDAWLAELAALPLVHQPGERVTYS-HAIDLLG-VIMSRIDDKPFYQV
MLBr_00119|M.leprae_Br4923 ---HAYAQQKLSELRPFYRPGLVHIYHALTWGPLMREIIWTATGKEIRDI
*. * :: .
Mflv_0495|M.gilvum_PYR-GCK VDRNVFAPLKMSETRYLP----VTKVCGPHRVRGTAIAADPH--GQAVDN
MSMEG_6805|M.smegmatis_MC2_155 VREHVFAPLGMSDTYYLP----AANACGPHEIRGNALVSDPD--GPRTTD
MAB_4800|M.abscessus_ATCC_1997 VQDNIFAPLGMNETRYLP----AAKACGPHHIVGTAITLEEG--PRAAPD
Mvan_5895|M.vanbaalenii_PYR-1 VRDRVFAPLGMTDTRYLP----AAKTCGPHEIRGTTVALGHY--PPWIGQ
TH_4016|M.thermoresistible__bu VTENVFAPLGMADTRYLP----VSKACGPIRMVGNAVVEDES--AEPTP-
MMAR_3399|M.marinum_M VAQHVFAPIGMQDSDFLP----AAKACGPHSVRGAAIGWAPAPAGRPRAP
MAV_3991|M.avium_104 VQHHVFGPLGMHDTRYLP----PAKACGPHIIRGAAIAWAPN--GEPMTE
Mb1759c|M.bovis_AF2122/97 VQRLLLDPLQLAHTRYFSDQIIGLNVAASHSVVDGKPIAVTDFWTFPRSC
Rv1730c|M.tuberculosis_H37Rv VQRLLLDPLQLAHTRYFSDQIIGLNVAASHSVVDGKPIAVTDFWTFPRSC
MUL_1772|M.ulcerans_Agy99 LDERILGPAGMTDTGFFV--------------------------STQAQR
MLBr_00119|M.leprae_Br4923 LAAEILDPLGFRWTNFGVA-------------------------EEDVPL
: :: * : : :
Mflv_0495|M.gilvum_PYR-GCK CPDGTWNIDLLERIAPTALDEDT---PGLNPDYG-YPLRGTVHDPTARRM
MSMEG_6805|M.smegmatis_MC2_155 CPADSWSTGLLTRVAPTALDEDT---PGINPHFG-LPLRGTVHDPTARRM
MAB_4800|M.abscessus_ATCC_1997 CATGAWSTALLTRIAPTAHDEDT---PGINPHYG-RLIRGTVHDPTARRM
Mvan_5895|M.vanbaalenii_PYR-1 CPAGTWSTALLPRIAPTARDGDT---PGLNPDYG-HLLRGTVHDPTARRM
TH_4016|M.thermoresistible__bu CGEDRWSTSLRARIAPTSCDGDT---PDLNPHRG-RLLRGTVHDPTARRM
MMAR_3399|M.marinum_M CPAGTWDTALLSHIAPTALDEESGADPSRNPNFN-RLLRGTVHDPTARRM
MAV_3991|M.avium_104 CPEWTWRTTLLPRIAPTARDEESRDDPSKNPDYY-ALLRGTVHDPTARRM
Mb1759c|M.bovis_AF2122/97 NPTGGLMSTARDQLRYAQFHLGDGRAPNGEQILSRQSLKAMRSNPGAGGT
Rv1730c|M.tuberculosis_H37Rv NPTGGLMSTARDQLRYAQFHLGDGRAPNGEQILSRQSLKAMRSNPGAGGT
MUL_1772|M.ulcerans_Agy99 RAATMYRLDELDQLR-----------------------HDVMGPPHVRPP
MLBr_00119|M.leprae_Br4923 VALSHATGRPLPPIIAAIFR----------KAIGGTVHEVIPYTNTPPFL
: .
Mflv_0495|M.gilvum_PYR-GCK GGVAGSAGVFTTVHDMGRFAQALLDR---VAGRPSPFPLSQAGARMMTTP
MSMEG_6805|M.smegmatis_MC2_155 GGVAGSAGVFTTVHDLGLFAQALLDR---RANRPSTFPLQQATVELMTTP
MAB_4800|M.abscessus_ATCC_1997 GGVAGSAGVFSTAGDVGRYCQALLDR---LAGRPSTFPLKRATLELMTSP
Mvan_5895|M.vanbaalenii_PYR-1 GGVTGSAGVFSTVGDVGRFAQALLDG---LAGRPSAFPLTRSTLEMMTTP
TH_4016|M.thermoresistible__bu GGVSGSAGVFSTAADVGSYAQALLDR---LAGRPSRFPLRRSTLQMMTSP
MMAR_3399|M.marinum_M GGVAGHAGVFSTAHDVSSFAQALLDR---LAGRPSTFPLTQATCELMTTP
MAV_3991|M.avium_104 GGVAGNAGVFSTAHDVGVYAQALLDR---LAGRPSDFPLQQGTVELMTTP
Mb1759c|M.bovis_AF2122/97 LWVELTGMGVTWMLRPSAENVTIVEHGGTWKGQRSGFVMVPDRNFAMTVL
Rv1730c|M.tuberculosis_H37Rv LWVELTGMGVTWMLRPSAENVTIVEHGGTWKGQRSGFVMVPDRNFAMTVL
MUL_1772|M.ulcerans_Agy99 SFCNAGGGLWSTADDYLRFVRLLLGD----GTIDGVRVLSPESVRLMRTD
MLBr_00119|M.leprae_Br4923 TTIVPSSNTVSTANELSRFAEIWLRG----GELDGIRVLRSETLLGAVAE
. : : . :
Mflv_0495|M.gilvum_PYR-GCK QQPA-----------------------------------PGRDLRG----
MSMEG_6805|M.smegmatis_MC2_155 QQPG-----------------------------------AGPDLRG----
MAB_4800|M.abscessus_ATCC_1997 AQPGHNDGQLKAANDAARLAIEKTSNSTDPLLAPGYPAIAGQNLRG----
Mvan_5895|M.vanbaalenii_PYR-1 QQPGHTPGQLEAANDAIRTATENDPDST----ARNYPAVPGQDLRG----
TH_4016|M.thermoresistible__bu QQPGHAPGQLDVANKAAREAVLSASDDP--LLAPDYPAVPGRDLRG----
MMAR_3399|M.marinum_M QQPGHDMAQLDAANAAAREALAHRPNRTDPMLAPRYPAISGQTLFG----
MAV_3991|M.avium_104 QQPGHDPRQLEAANRAARSG------------TPNYPAIKGQNLFG----
Mb1759c|M.bovis_AF2122/97 TNSDGGFHMINDLFASDWALQRFAGLSNLPATPQRLGAVDLAPYEGRYIA
Rv1730c|M.tuberculosis_H37Rv TNSDGGFHMINDLFASDWALQRFAGLSNLPATPQRLGAVDLAPYEGRYIA
MUL_1772|M.ulcerans_Agy99 RLSDEHKR-------------------------HNFLGAPFWVGRG----
MLBr_00119|M.leprae_Br4923 CRRLR----------------------------PDFVVGLMPARWG----
*
Mflv_0495|M.gilvum_PYR-GCK ----FGWDIDTAHSGPRGS------VFPVGSFGHTGFTGVS-VWLDPESD
MSMEG_6805|M.smegmatis_MC2_155 ----LGWDIDTPHSRPRGT------VFPVGSFGHTGFTGVS-MWMDPGSD
MAB_4800|M.abscessus_ATCC_1997 ----LGWDIDTELSKPRGM------LFPIGSFGHSGFTGVT-LWLDPGSD
Mvan_5895|M.vanbaalenii_PYR-1 ----FGWDIDTAHSRPRGT------LFPVGSFGHAGFTGVT-LWMDPGSD
TH_4016|M.thermoresistible__bu ----FGWDIDTNQSRPRGS------VFPVGSFGHTGFTGVT-LWIDPGSD
MMAR_3399|M.marinum_M ----FGWDIDTPYSAPRGV------VFPIGSFGGTGFTGTS-LWIDPGSD
MAV_3991|M.avium_104 ----FGWDIDTGYSRPRGR------VFPIGSFGNTGFTGTT-LWMDPGSD
Mb1759c|M.bovis_AF2122/97 KQVAQNGDLETTVIDFRARDGQLAGSMSTDDANPDGQNSAN-LGLAFYRP
Rv1730c|M.tuberculosis_H37Rv KQVAQNGDLETTVIDFRARDGQLAGSMSTDDANPDGQNSAN-LGLAFYRP
MUL_1772|M.ulcerans_Agy99 ----FGLNLSVVTDPVQSTP-----LFGPGGLGTFSWPGAYGTWWQADPG
MLBr_00119|M.leprae_Br4923 ----TGFILGTNKFGPFGRN-------APAAFGHLGLVNIA-IWADPQRS
. : . . . . .
Mflv_0495|M.gilvum_PYR-GCK TYVVILGNVIHQRGGPP--ITPLSGA--VATEAARALHLYGS-------
MSMEG_6805|M.smegmatis_MC2_155 TYVIVLANVIHQRGGPP--IATLSGD--VATEAARALHLYGT-------
MAB_4800|M.abscessus_ATCC_1997 TYVVVLANVIHQRGGPP--IVRLSGH--IATVAARMLDLYRN-------
Mvan_5895|M.vanbaalenii_PYR-1 TYVVVLANVIHQPGGPP--IVNLSGE--VATATAQALGLYR--------
TH_4016|M.thermoresistible__bu TYVVVLANVIHQPGGPP--VAPLSGR--VATLAARALHLYGT-------
MMAR_3399|M.marinum_M SYVVLLSNAIHLRGSTP--ISDLRGQ--VATAAALALGL----------
MAV_3991|M.avium_104 TYVILLSNSIHLRGSPP--ISNLRGD--VATAAARALHLYGAAPP----
Mb1759c|M.bovis_AF2122/97 DYGLDLGPDNKPTGSRSNFVRGPDGN--IAWFCSQHGRLFRRQ------
Rv1730c|M.tuberculosis_H37Rv DYGLDLGPDNKPTGSRSNFVRGPDGN--IAWFCSQHGRLFRRQ------
MUL_1772|M.ulcerans_Agy99 ADLILLYLIQHCPDLSVNAAAAVAGNPGLAKLRTAQPRFVRRTYRALGL
MLBr_00119|M.leprae_Br4923 MAAGLISS--GKPGQDP--NAGRYGA--LMNTITAEIPVH---------
: . * : : .