For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAPVNVISVAVVASDPLTRDGALARLSSHRELDVRAWQAGCETSVLLVLATTITAPLLCQIEDVQKDGPS HAPKLVVVADEFSAEQVFRMIKLGLTGLLYRSQSTFDCIVETIRLSAEGRLRLPERVQRYLVGRIKSTPT AEPDTPCAAALAEREVAVLRLLADGLSTHQVAVQLNYCERTIKNIVHDIVTRLKLRNRTHAVAHALRAGL I
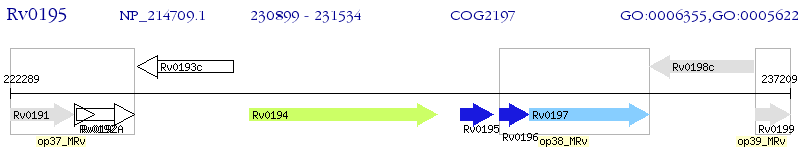
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0195 | - | - | 100% (211) | POSSIBLE TWO COMPONENT TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY LUXR-FAMILY) |
| M. tuberculosis H37Rv | Rv3133c | devR | 5e-08 | 25.36% (209) | two component transcriptional regulatory protein DevR |
| M. tuberculosis H37Rv | Rv0844c | narL | 4e-06 | 25.23% (214) | nitrate/nitrite response transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0201 | - | 1e-116 | 100.00% (211) | LuxR family two component transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3579 | - | 6e-12 | 26.95% (141) | two component LuxR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4519c | - | 8e-16 | 30.88% (217) | two-component LuxR family response regulator |
| M. marinum M | MMAR_3959 | - | 5e-13 | 31.25% (144) | two-component transcriptional regulatory protein |
| M. avium 104 | MAV_4114 | - | 9e-12 | 26.95% (141) | response regulator receiver domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3240 | - | 1e-12 | 30.00% (140) | DNA-binding response regulator, LuxR family protein |
| M. thermoresistible (build 8) | TH_3563 | - | 8e-14 | 31.43% (140) | PUTATIVE DNA-binding response regulator, LuxR family protein |
| M. ulcerans Agy99 | MUL_3814 | - | 1e-13 | 31.94% (144) | two component transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_2836 | - | 8e-13 | 27.96% (211) | two component LuxR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3579|M.gilvum_PYR-GCK -M--GIDRT-SVMVVDDHPIWRD-AVARDLADDGFDVVATADGVASAKRR
Mvan_2836|M.vanbaalenii_PYR-1 -----------MMVVDDHPIWRD-AVARDLADDGFDVVATADGVASAKRR
MSMEG_3240|M.smegmatis_MC2_155 MM--TDAAP-TVMVVDDHPIWRD-AVARDLADDGFDVVATADGVASASRR
TH_3563|M.thermoresistible__bu MG--EPQQP-TVMVVDDHPIWRE-AVARDLAEAGLSVVATADGVAAARRR
MMAR_3959|M.marinum_M MS--EHESP-TVMVVDDHPIWRD-AVARDLSDDGFAVVATADGVATAQRR
MUL_3814|M.ulcerans_Agy99 MS--EHESP-TVMVVDDHPIWRD-AVARDLSDDGFAVVATADGVATAQRR
MAV_4114|M.avium_104 MT--DDATPTTVMVVDDHPIWRD-AVARDLAESGFAVVATADGVATAQRR
MAB_4519c|M.abscessus_ATCC_199 MTGNTDSEQISVMVVDDHPMWRD-GVSRDLQAAGLRVVATADGVASSGRI
Rv0195|M.tuberculosis_H37Rv ---MAPVNVISVAVVASDPLTRDGALARLSSHRELDVRAWQAGCETS---
Mb0201|M.bovis_AF2122/97 ---MAPVNVISVAVVASDPLTRDGALARLSSHRELDVRAWQAGCETS---
: ** ..*: *: .::* : * * * ::
Mflv_3579|M.gilvum_PYR-GCK AAVVRPDVVVMDMRLGDGDGAQATTEVLAVSPSSRILVLSASDERDDVLE
Mvan_2836|M.vanbaalenii_PYR-1 AAVVQPAVVLMDMRLADGDGAQATAEVLAVSPSSRVLVLSASDERDDVLE
MSMEG_3240|M.smegmatis_MC2_155 AAVVRPDVVLMDMRLGDGSGAQATAEVLAVSPRSRVLVLSASDERDDVLQ
TH_3563|M.thermoresistible__bu AAAVRPDVVLMDMRLPDGDGARATAEVRAVSPDSRVLVLSASEERDDVLE
MMAR_3959|M.marinum_M AAIVKPDVVVMDMRLADGDGVQATTEVLKVSPATRVLVLSASDEREDVLE
MUL_3814|M.ulcerans_Agy99 AAIVKPDVVVMDMRLADGDGVQATTEVLKVSPATRVLVLSTSDEREDVLE
MAV_4114|M.avium_104 AGVVRPDVVVMDMQLPDGDGAQATAAVLAVSPSSRVLVLSASDERDDVLQ
MAB_4519c|M.abscessus_ATCC_199 AATVKPDVVLMDMQLTDGDGAQATAAVLSASPHSHILVLSASAEREDVLE
Rv0195|M.tuberculosis_H37Rv -VLLVLATTITAPLLCQIEDVQKD----GPSHAPKLVVVADEFSAEQVFR
Mb0201|M.bovis_AF2122/97 -VLLVLATTITAPLLCQIEDVQKD----GPSHAPKLVVVADEFSAEQVFR
: ..: * : ...: * .:::*:: . . ::*:.
Mflv_3579|M.gilvum_PYR-GCK AVKAGATGYLVKSASKQE-LGDAVRATAQGRAVFTPGLAGLVLGEFRRIE
Mvan_2836|M.vanbaalenii_PYR-1 AVKAGATGYLVKSASKQE-LGDAVRATAQGRAVFTPGLAGLVLGEFRRIE
MSMEG_3240|M.smegmatis_MC2_155 AVKAGATGYLVKSASRTE-LADAVRATAEGRAVFTPGLAGLVLGEYRRIA
TH_3563|M.thermoresistible__bu AVKAGATGYLVKSASKTE-LVDAVHATAQGRAVFTPGLAGLVLGEYRRIS
MMAR_3959|M.marinum_M AVKAGATGYLVKSASKAE-LADAVRATAAGRAVFTPSLAGLVLGEYRRIA
MUL_3814|M.ulcerans_Agy99 AVKTGATGYLVKSASKAE-LADAVRATAAGRAVFTPSLAGLVLGEYRRIA
MAV_4114|M.avium_104 AVKAGAAGYLVKSASKAE-LAQAVTDTAAGRAVFTPSLAGLVLGEYRRIA
MAB_4519c|M.abscessus_ATCC_199 AVKAGATGYLVKSASAAE-LIDAVRATAKGQAVFTPGLAGLVLGEFRRMS
Rv0195|M.tuberculosis_H37Rv MIKLGLTGLLYRSQSTFDCIVETIRLSAEGRLRLPERVQRYLVGRIKSTP
Mb0201|M.bovis_AF2122/97 MIKLGLTGLLYRSQSTFDCIVETIRLSAEGRLRLPERVQRYLVGRIKSTP
:* * :* * :* * : : ::: :* *: :. : ::*. :
Mflv_3579|M.gilvum_PYR-GCK QS-SPAGADGPTLTERETEILRYVAKGLTAKQIASRLSLSHRTVENHVQA
Mvan_2836|M.vanbaalenii_PYR-1 RD-SPAG---PTLTERETEILRHVAKGLTAKQIASRLSLSHRTVENHVQA
MSMEG_3240|M.smegmatis_MC2_155 QQPAQEGPATPTLTERETEILRYVAKGLTAKQIAARLSLSHRTVENHVQA
TH_3563|M.thermoresistible__bu RQ-PGAGPDAPRLTERETEILRYVAKGLTAKQIAAQLSLSHRTVENHVQA
MMAR_3959|M.marinum_M QT-ADSSQDRPSLTERETEILRYVAKGLSAKQIAEKLSLSHRTVENHVQA
MUL_3814|M.ulcerans_Agy99 QT-ADSSQDRPSLTERETEILRYVAKGLSAKQIAEKLSLSHRTVENHVQA
MAV_4114|M.avium_104 QS-KDDGPTRPRLTDRETEVLRYVAKGLSAKQIAEKLSLSHRTVENHVQA
MAB_4519c|M.abscessus_ATCC_199 SAPAADADETPRLSDRETEVLRLVAKGLSAKQIATRLSLSHRTVENHVQS
Rv0195|M.tuberculosis_H37Rv TA-EPDTPCAAALAEREVAVLRLLADGLSTHQVAVQLNYCERTIKNIVHD
Mb0201|M.bovis_AF2122/97 TA-EPDTPCAAALAEREVAVLRLLADGLSTHQVAVQLNYCERTIKNIVHD
. *::**. :** :*.**:::*:* :*. ..**::* *:
Mflv_3579|M.gilvum_PYR-GCK TFRKLQVGNRVELARYAIEHGLDE--
Mvan_2836|M.vanbaalenii_PYR-1 TFRKLQIGNRVELARYAIEHGLDE--
MSMEG_3240|M.smegmatis_MC2_155 TFRKLQVANRVELARYAIEHGLDE--
TH_3563|M.thermoresistible__bu TFRKLQVANRVELARYAIEHGLDE--
MMAR_3959|M.marinum_M TFRKLQVANRVELTRYAIEHGLDK--
MUL_3814|M.ulcerans_Agy99 TFRKLQLANRVELTRYAIEHGLDK--
MAV_4114|M.avium_104 TFRKLQVANRVELARYAIEHGLDEEP
MAB_4519c|M.abscessus_ATCC_199 TLRKLQLGNRVELTRYAIEHGLDE--
Rv0195|M.tuberculosis_H37Rv IVTRLKLRNRTHAVAHALRAGLI---
Mb0201|M.bovis_AF2122/97 IVTRLKLRNRTHAVAHALRAGLI---
. :*:: **.. . :*:. **