For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSCDSGLCTTPFGFSSTAADVLAGIDLSGRRAIITGATSGIGVETARALAAAGADVVLGVRRLDAGRRV AAQIVEDTGNGAVTAAQVDVADLNSVREFVSRFSGAPVHMLINNAGVMALPELSRTTDGREMQFATNYLG HFALTVGLYPALEAACGARVVSVSSSGHLLSPVVFDDIDYRFRPYDPWTAYGQSKTADILLAVGVTQRWG DAGIVGNALHPGAVATGLQKHTGGLRTPADRRKTVAQGAATSVLLAASPLLEGVGGRYFEDCNESPVVAE RPDDFSGVAQYALDPDNADRLWDGQRSFSP
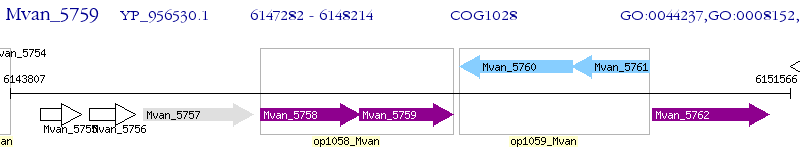
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5759 | - | - | 100% (310) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_0945 | - | 8e-58 | 44.34% (318) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4584 | - | 4e-53 | 41.30% (322) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2286 | - | 2e-40 | 45.37% (216) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2120 | - | 2e-52 | 41.83% (306) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2263 | - | 2e-40 | 45.37% (216) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 2e-35 | 40.00% (260) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0646c | - | 2e-37 | 41.60% (250) | short chain dehydrogenase |
| M. marinum M | MMAR_3355 | - | 3e-39 | 43.52% (216) | oxidoreductase |
| M. avium 104 | MAV_4710 | - | 3e-38 | 42.17% (249) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0876 | - | 1e-38 | 41.13% (248) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3199 | - | 1e-37 | 43.58% (218) | PUTATIVE retinol dehydrogenase 13 |
| M. ulcerans Agy99 | MUL_1288 | - | 1e-39 | 43.52% (216) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0646c|M.abscessus_ATCC_199 ------------MSRSTTKWSTTDIPDQTGRIAIVTGANTGLGLETAKAL
MSMEG_0876|M.smegmatis_MC2_155 ----------------MTKWTTARIPDQTGRVAIVTGANTGLGLETAKAL
MLBr_00315|M.leprae_Br4923 ----------------MAKWTTADIPDQTGRVAVITGANTGLGYQTALAL
MAV_4710|M.avium_104 ---------MAPHDKRHRDWSEADVGDQSGRVVVITGANTGIGYETAAVL
Mb2286|M.bovis_AF2122/97 ----------------MAKDLVATVPDLSGKLAIITGANSGLGFGLARRL
Rv2263|M.tuberculosis_H37Rv ----------------MAKDLVATVPDLSGKLAIITGANSGLGFGLARRL
MMAR_3355|M.marinum_M ----------------MASDLIATVPDLSGKLAIVTGANSGLGFGLARRL
MUL_1288|M.ulcerans_Agy99 ----------------MASDLIATVPDLSGKLAIVTGANSGLGFGLARRL
TH_3199|M.thermoresistible__bu ------------------MWTAADLPSFSDRTVIVTGANSGLGLVTAREL
Mvan_5759|M.vanbaalenii_PYR-1 MSSCDSGLCTTPFGFSSTAADVLAGIDLSGRRAIITGATSGIGVETARAL
Mflv_2120|M.gilvum_PYR-GCK MTTTS----------ETTALDIVAGADLGGKTCVITGASSGLGRESARAL
. .: ::***.:*:* * *
MAB_0646c|M.abscessus_ATCC_199 AAHGAHVVLAVRNAEKGKAAAEAITAAHSNADVTLQSLDLSSLESVRRAS
MSMEG_0876|M.smegmatis_MC2_155 AAKGAHVVLAVRNLDKGKAAVDWIARSAPTADLELQQLDLGSLASVRAAA
MLBr_00315|M.leprae_Br4923 AEHGAHVVLAVRNLDKGKDAAARITATSAQNNVALQELDLASLESVRAAA
MAV_4710|M.avium_104 AHRGAHVVLAVRDLEKGNAALSRIVAASPNADVTLQQLDLASLASVRSAA
Mb2286|M.bovis_AF2122/97 SAAGADVIMAIRNRAKGEAAVEEIRTAVPDAKLTIKALDLSSLASVAALG
Rv2263|M.tuberculosis_H37Rv SAAGADVIMAIRNRAKGEAAVEEIRTAVPDAKLTIKALDLSSLASVAALG
MMAR_3355|M.marinum_M SAAGADVVMAIRNRAKGEAAIEEIRSAVPDAKLSIKALDLSSLASVAALG
MUL_1288|M.ulcerans_Agy99 SAAGADVVMAIRNRAKGEAAIEEIRSAVPDAKLSTKALDLSSLASVAALG
TH_3199|M.thermoresistible__bu ARVGARVIIAVRNTTKGEAAAEQMTGPG-HGPVEVRALDLQNLASVREFA
Mvan_5759|M.vanbaalenii_PYR-1 AAAGADVVLGVRRLDAGRRVAAQIVEDTGNGAVTAAQVDVADLNSVREFV
Mflv_2120|M.gilvum_PYR-GCK ASTGAHVILAARNLEALSDTEAWVRAEVADAELSIVPLDLTSLASVASAA
: ** *::. * . : : :*: .* **
MAB_0646c|M.abscessus_ATCC_199 DELKGRYDKIDLLINNAGVMWTE-KSSTADGFELQFGTNHLGHYALTGLL
MSMEG_0876|M.smegmatis_MC2_155 DDLKGKFDRIDLLVNNAGVMWPP-RQTTADGFELQFGTNHLGHFALTGLL
MLBr_00315|M.leprae_Br4923 KQLRSDYDHIDLLINNAGVMWTP-KSTTKDGFELQFGTNHLGHFAFTGLL
MAV_4710|M.avium_104 EALRAAYPRIDLLINNAGVMWTP-KQVTEDGFELQFGTNHLGHFALTGLL
Mb2286|M.bovis_AF2122/97 EQLMADGRPIDLLINNAGVMTPPERVTTADGFELQFGSNHLGHFALTAHL
Rv2263|M.tuberculosis_H37Rv EQLMADGRPIDLLINNAGVMTPPERVTTADGFELQFGSNHLGHFALTAHL
MMAR_3355|M.marinum_M DQLNSEGRPIDILINNAGVMTPPERDTTADGFELQFGSNHLGHFALTAHV
MUL_1288|M.ulcerans_Agy99 DQLNSEGRPIDILINNAGVMTPPERDTTADGFELQFGSNHLGHFALTAHV
TH_3199|M.thermoresistible__bu AGVEG---DVDVLVNNAGIMAVP-LARTVDGFESQIGTNHLGHFALTNLL
Mvan_5759|M.vanbaalenii_PYR-1 SRFSGAP--VHMLINNAGVMALPELSRTTDGREMQFATNYLGHFALTVGL
Mflv_2120|M.gilvum_PYR-GCK AQISELTPAVHVLMNNAGVMFTP-FGRTAEGFETQFGTNHLGHFEFTRLL
. :.:*:****:* * :* * *:.:*:***: :* :
MAB_0646c|M.abscessus_ATCC_199 LERLLPVEGSRVVTVSSIGHRIRADIHFDDLQWERD-YDRVAAYGQSKLA
MSMEG_0876|M.smegmatis_MC2_155 LDRMLTVPGSRVVTVSSQGHRILAAIHFDDLQWERR-YNRVAAYGQSKLA
MLBr_00315|M.leprae_Br4923 LDRLLPIVGSRVITVSSLSHRLFADIHFNDLQWECN-YNRVAAYGQSKLA
MAV_4710|M.avium_104 LDHLLGVRDSRVVTVSSLGHRLRAAIHFDDLHWERR-YDRVAAYGQSKLA
Mb2286|M.bovis_AF2122/97 LPLLRAAQRARVVSLSSLAAR-RGRIHFDDLQFERS-YAPMTAYGQSKLA
Rv2263|M.tuberculosis_H37Rv LPLLRAAQRARVVSLSSLAAR-RGRIHFDDLQFERS-YAPMTAYGQSKLA
MMAR_3355|M.marinum_M LPLLRAAQGARVVSLSSLAAR-RGRIHFDDLQFEKS-YAAMTAYGQSKLA
MUL_1288|M.ulcerans_Agy99 LPLLRAAQGARVVSLSSLAAR-RGRIHFDDLQFEKS-YAAMTAYGQSKLA
TH_3199|M.thermoresistible__bu LPQIT----DRVVTVSSLMHW-FGYLSLNDLNWKARPYSAWLAYAQSKLA
Mvan_5759|M.vanbaalenii_PYR-1 YPALEAACGARVVSVSSSGHL-LSPVVFDDIDYRFRPYDPWTAYGQSKTA
Mflv_2120|M.gilvum_PYR-GCK FPALVAADGARVVNLSSEGHR-ISDVDFEDPNWETRDYDKFAAYGASKTA
: **:.:** . : ::* .:. * **. ** *
MAB_0646c|M.abscessus_ATCC_199 NLLFTYELQRRLAG---TNTVALAAHPGGSNTEL-------ARNSPLWVR
MSMEG_0876|M.smegmatis_MC2_155 NLLFTYELQRRLTG---HQTTALAAHPGASNTEL-------ARHLP----
MLBr_00315|M.leprae_Br4923 NLLFTYELQRRLATR--QTTIAVAAHPGGSRTEL-------TRTLPALIA
MAV_4710|M.avium_104 NLLFTYELQRRLAAAPDAKTIAVAAHPGGSNTEL-------ARHLPGIFR
Mb2286|M.bovis_AF2122/97 VLMFARELDRRSRAAG-WGIISNAAHPGLTKTNLQIAGPSHGRDKPALME
Rv2263|M.tuberculosis_H37Rv VLMFARELDRRSRAAG-WGIISNAAHPGLTKTNLQIAGPSHGRDKPALME
MMAR_3355|M.marinum_M VLMFARELDRRSRAAG-WGVMSNAAHPGLTKTNLQISGPSHGREKPALMQ
MUL_1288|M.ulcerans_Agy99 VLMFARELDRRSRAAG-WGVMSNAAHPGLTKTNLQISGPSHGREKPALMQ
TH_3199|M.thermoresistible__bu NLLFTSELQRRLDTVG-SPVRAVAAHPGYSSTNLQG---HTGRRFGDALA
Mvan_5759|M.vanbaalenii_PYR-1 DILLAVGVTQRWGDAG---IVGNALHPGAVATGLQK------HTG-----
Mflv_2120|M.gilvum_PYR-GCK NVLHAVELDRRLRDSG---VRAFAVHPGIVATSLAR------HMTNDDFA
:: : : :* . * *** * * :
MAB_0646c|M.abscessus_ATCC_199 AVFDV---VAPVLVQGADMGALPTLRAATDPAALGGQYYGPDGFMEQRGN
MSMEG_0876|M.smegmatis_MC2_155 GALER---LVTPLAQDAALGALPTLRAATDPGALGGQYFGPDGIGETRGY
MLBr_00315|M.leprae_Br4923 PIFSV---AELFLTQDAATGALPTLRAATDAAVLGGQYFGPDGFAEIRGH
MAV_4710|M.avium_104 PVQAV---LGPVLFQSPAMGALPTLRAATDPAVQGAQYYGPDGFLEQRGR
Mb2286|M.bovis_AF2122/97 RLYKTSWRFAPFLWQEIEEGILPALYAAATPQADGGAFYGPRGRYEVAGG
Rv2263|M.tuberculosis_H37Rv RLYKTSWRFAPFLWQEIEEGILPALYAAATPQADGGAFYGPRGRYEVAGG
MMAR_3355|M.marinum_M RFYTTSWRFAPFLWQEIEDGILPALYAAVTPQAEGGAFYGPRGFYEAAGG
MUL_1288|M.ulcerans_Agy99 RFYTTSWRFAPFLWQEIEDGILPALYAAVTPQAEGGAFYGPRGFYEAAGG
TH_3199|M.thermoresistible__bu RIGNR------YLATDAEFGARQTLYAVAR-DVPGDSFIGPR--FAMHGP
Mvan_5759|M.vanbaalenii_PYR-1 ------GLRTP----ADRRK---TVAQGAATSVLLAASPLLEGVGGRYFE
Mflv_2120|M.gilvum_PYR-GCK SLNKSTASRNPDKPTTDFRKQFTTPEHGAATQVWAAVSDDLDGQGGLYLS
: . .
MAB_0646c|M.abscessus_ATCC_199 -PKVVASSEQSYNLDLQRRLWSVSEELTDVVFPVK------
MSMEG_0876|M.smegmatis_MC2_155 -PKVVASSAQSHDADLQRRLWAVSEELTGVTFPVGEALQQA
MLBr_00315|M.leprae_Br4923 -PKVVASNGKSHDVDRQLRLWAVSEELTGVVYPVG------
MAV_4710|M.avium_104 -PKLVESSAQSHDEQLQRRLWAVSEELTGVHFPV-------
Mb2286|M.bovis_AF2122/97 GVREAKVPAAARNDADSKRLWEVSEQLTGVSYPKSR-----
Rv2263|M.tuberculosis_H37Rv GVREAKVPAAARNDADSKRLWEVSEQLTGVSYPKSR-----
MMAR_3355|M.marinum_M GVRAAKVPARAGNDADCQRLWEVSERLTGVGYPTSS-----
MUL_1288|M.ulcerans_Agy99 GVRAAKVPARAGNDADCQRLWEVSERLTGVGYPTSS-----
TH_3199|M.thermoresistible__bu -TEQVGRSPLARSAGTARALWELSEQLTDTKFPL-------
Mvan_5759|M.vanbaalenii_PYR-1 DCNESPVVAERPDDFSGVAQYALDPDNADRLWDGQRSFSP-
Mflv_2120|M.gilvum_PYR-GCK DCRIREAAPYAADETRALTLWALS----EHLCTASSAT---
. . : :.