For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TNKQQPLGSGFTAASTAADVLAGIDLTGRNAIVTAGHVGLGLETTRALADAGANVVVASRNPETAAAALV GVERVRVEQLDLMDPASIERFVAGYGEAALHILINNAGIMGGDLVRDARGYEAQFATNHLGHFQLTNGLL PALRAADGARVVEVSSWGHHLSDIRWEDPHFESDYDGMVAYGQSKTANVLFAVELDRRWADDGVRGYSLH PGGIVSTNLGPSFTLDDWRAMGLVDDGGQPIVDPDRDMKTPQQGASTQVFAATSPQLADIGGVYLANNDI APLEADATPVTVDMGAGPLQTTPGVTGYAVDPESAERLWELSEKLLA*
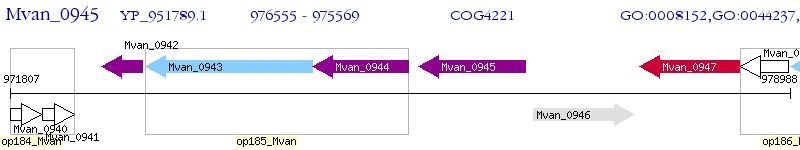
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0945 | - | - | 100% (328) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4584 | - | 1e-62 | 44.17% (326) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_5759 | - | 8e-58 | 44.34% (318) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2286 | - | 1e-33 | 35.69% (339) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2120 | - | 7e-61 | 43.56% (326) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2263 | - | 1e-33 | 35.69% (339) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 4e-31 | 41.35% (208) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0646c | - | 1e-39 | 38.46% (312) | short chain dehydrogenase |
| M. marinum M | MMAR_3355 | - | 2e-34 | 37.35% (340) | oxidoreductase |
| M. avium 104 | MAV_4710 | - | 8e-33 | 35.78% (313) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0876 | - | 1e-33 | 35.81% (310) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4179 | - | 2e-34 | 35.65% (331) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1288 | - | 3e-34 | 37.35% (340) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_4710|M.avium_104 --------MAPHDKRHRDWSEADVGDQSGRVVVITGANTGIGYETAAVLA
TH_4179|M.thermoresistible__bu --------------MTGKWTARDVPDQTGRTAVITGANTGLGFETAKVLA
MAB_0646c|M.abscessus_ATCC_199 -----------MSRSTTKWSTTDIPDQTGRIAIVTGANTGLGLETAKALA
MSMEG_0876|M.smegmatis_MC2_155 ---------------MTKWTTARIPDQTGRVAIVTGANTGLGLETAKALA
MLBr_00315|M.leprae_Br4923 ---------------MAKWTTADIPDQTGRVAVITGANTGLGYQTALALA
Mb2286|M.bovis_AF2122/97 ---------------MAKDLVATVPDLSGKLAIITGANSGLGFGLARRLS
Rv2263|M.tuberculosis_H37Rv ---------------MAKDLVATVPDLSGKLAIITGANSGLGFGLARRLS
MMAR_3355|M.marinum_M ---------------MASDLIATVPDLSGKLAIVTGANSGLGFGLARRLS
MUL_1288|M.ulcerans_Agy99 ---------------MASDLIATVPDLSGKLAIVTGANSGLGFGLARRLS
Mvan_0945|M.vanbaalenii_PYR-1 MTNKQQPLGSGFTAASTAADVLAGIDLTGRNAIVTAGHVGLGLETTRALA
Mflv_2120|M.gilvum_PYR-GCK MTT---------TSETTALDIVAGADLGGKTCVITGASSGLGRESARALA
* *: ::*.. *:* : *:
MAV_4710|M.avium_104 HRGAHVVLAVRDLEKGNAALSRIVAASPNADVTLQQLDLASLASVRSAAE
TH_4179|M.thermoresistible__bu EKGAHVVLAVRDPDKGRRAADRITAAAPHADVTVRQLDLTSLDNIRRAAD
MAB_0646c|M.abscessus_ATCC_199 AHGAHVVLAVRNAEKGKAAAEAITAAHSNADVTLQSLDLSSLESVRRASD
MSMEG_0876|M.smegmatis_MC2_155 AKGAHVVLAVRNLDKGKAAVDWIARSAPTADLELQQLDLGSLASVRAAAD
MLBr_00315|M.leprae_Br4923 EHGAHVVLAVRNLDKGKDAAARITATSAQNNVALQELDLASLESVRAAAK
Mb2286|M.bovis_AF2122/97 AAGADVIMAIRNRAKGEAAVEEIRTAVPDAKLTIKALDLSSLASVAALGE
Rv2263|M.tuberculosis_H37Rv AAGADVIMAIRNRAKGEAAVEEIRTAVPDAKLTIKALDLSSLASVAALGE
MMAR_3355|M.marinum_M AAGADVVMAIRNRAKGEAAIEEIRSAVPDAKLSIKALDLSSLASVAALGD
MUL_1288|M.ulcerans_Agy99 AAGADVVMAIRNRAKGEAAIEEIRSAVPDAKLSTKALDLSSLASVAALGD
Mvan_0945|M.vanbaalenii_PYR-1 DAGANVVVASRNPE--TAAAALVGVERVRVEQ----LDLMDPASIERFVA
Mflv_2120|M.gilvum_PYR-GCK STGAHVILAARNLEALSDTEAWVRAEVADAELSIVPLDLTSLASVASAAA
**.*::* *: : : . *** . .:
MAV_4710|M.avium_104 ALRAAYPRIDLLINNAGVMWTP-KQVTEDGFELQFGTNHLGHFALTGLLL
TH_4179|M.thermoresistible__bu DLRAGYPRIDLLINNAGVMYPP-RQTTRDGFELQFGTNHLGHFALTGQLL
MAB_0646c|M.abscessus_ATCC_199 ELKGRYDKIDLLINNAGVMWTE-KSSTADGFELQFGTNHLGHYALTGLLL
MSMEG_0876|M.smegmatis_MC2_155 DLKGKFDRIDLLVNNAGVMWPP-RQTTADGFELQFGTNHLGHFALTGLLL
MLBr_00315|M.leprae_Br4923 QLRSDYDHIDLLINNAGVMWTP-KSTTKDGFELQFGTNHLGHFAFTGLLL
Mb2286|M.bovis_AF2122/97 QLMADGRPIDLLINNAGVMTPPERVTTADGFELQFGSNHLGHFALTAHLL
Rv2263|M.tuberculosis_H37Rv QLMADGRPIDLLINNAGVMTPPERVTTADGFELQFGSNHLGHFALTAHLL
MMAR_3355|M.marinum_M QLNSEGRPIDILINNAGVMTPPERDTTADGFELQFGSNHLGHFALTAHVL
MUL_1288|M.ulcerans_Agy99 QLNSEGRPIDILINNAGVMTPPERDTTADGFELQFGSNHLGHFALTAHVL
Mvan_0945|M.vanbaalenii_PYR-1 GYGEA--ALHILINNAGIMGGD-LVRDARGYEAQFATNHLGHFQLTNGLL
Mflv_2120|M.gilvum_PYR-GCK QISELTPAVHVLMNNAGVMFTP-FGRTAEGFETQFGTNHLGHFEFTRLLF
:.:*:****:* *:* **.:*****: :* ::
MAV_4710|M.avium_104 DHLLGVRDSRVVTVSSLGHRLRAAIHFDDLHWER-RYDRVAAYGQSKLAN
TH_4179|M.thermoresistible__bu DNILPVDGSRVVTVASIAHRNMADIHFDDLQWER-GYHRVAAYGQSKLAN
MAB_0646c|M.abscessus_ATCC_199 ERLLPVEGSRVVTVSSIGHRIRADIHFDDLQWER-DYDRVAAYGQSKLAN
MSMEG_0876|M.smegmatis_MC2_155 DRMLTVPGSRVVTVSSQGHRILAAIHFDDLQWER-RYNRVAAYGQSKLAN
MLBr_00315|M.leprae_Br4923 DRLLPIVGSRVITVSSLSHRLFADIHFNDLQWEC-NYNRVAAYGQSKLAN
Mb2286|M.bovis_AF2122/97 PLLRAAQRARVVSLSSLAAR-RGRIHFDDLQFER-SYAPMTAYGQSKLAV
Rv2263|M.tuberculosis_H37Rv PLLRAAQRARVVSLSSLAAR-RGRIHFDDLQFER-SYAPMTAYGQSKLAV
MMAR_3355|M.marinum_M PLLRAAQGARVVSLSSLAAR-RGRIHFDDLQFEK-SYAAMTAYGQSKLAV
MUL_1288|M.ulcerans_Agy99 PLLRAAQGARVVSLSSLAAR-RGRIHFDDLQFEK-SYAAMTAYGQSKLAV
Mvan_0945|M.vanbaalenii_PYR-1 PALRAADGARVVEVSSWGHH-LSDIRWEDPHFES-DYDGMVAYGQSKTAN
Mflv_2120|M.gilvum_PYR-GCK PALVAADGARVVNLSSEGHR-ISDVDFEDPNWETRDYDKFAAYGASKTAN
: :**: ::* . : . : ::* ::* * ..*** ** *
MAV_4710|M.avium_104 LLFTYELQRRLAAAPDAKTIAVAAHPG-GSNTELARHLPGIFRPVQAVLG
TH_4179|M.thermoresistible__bu LMFAYELQRRLSAK-NAPTISVAAHPG-VSNTELTRYIPGARLPGVSLLA
MAB_0646c|M.abscessus_ATCC_199 LLFTYELQRRLAG---TNTVALAAHPG-GSNTELARNSPLWVRAVFDVVA
MSMEG_0876|M.smegmatis_MC2_155 LLFTYELQRRLTG---HQTTALAAHPG-ASNTELARHLP----GALERLV
MLBr_00315|M.leprae_Br4923 LLFTYELQRRLATR--QTTIAVAAHPG-GSRTELTRTLPALIAPIFSVAE
Mb2286|M.bovis_AF2122/97 LMFARELDRRSRAAG-WGIISNAAHPG-LTKTNLQIAGPSHGRDKPALME
Rv2263|M.tuberculosis_H37Rv LMFARELDRRSRAAG-WGIISNAAHPG-LTKTNLQIAGPSHGRDKPALME
MMAR_3355|M.marinum_M LMFARELDRRSRAAG-WGVMSNAAHPG-LTKTNLQISGPSHGREKPALMQ
MUL_1288|M.ulcerans_Agy99 LMFARELDRRSRAAG-WGVMSNAAHPG-LTKTNLQISGPSHGREKPALMQ
Mvan_0945|M.vanbaalenii_PYR-1 VLFAVELDRRWADDG---VRGYSLHPGGIVSTNLGPSFTLDDWRAMGLVD
Mflv_2120|M.gilvum_PYR-GCK VLHAVELDRRLRDSG---VRAFAVHPG-IVATSLARHMTNDDFASLNKST
::.: **:** . : *** *.* .
MAV_4710|M.avium_104 ------------PVLFQSPAMGALPTLRAATDPAVQ--GAQYYGPDGFLE
TH_4179|M.thermoresistible__bu ------------GLLTNSPAVGALATLRAATDPEVK--GGQYYGPDGFQE
MAB_0646c|M.abscessus_ATCC_199 ------------PVLVQGADMGALPTLRAATDPAAL--GGQYYGPDGFME
MSMEG_0876|M.smegmatis_MC2_155 ------------TPLAQDAALGALPTLRAATDPGAL--GGQYFGPDGIGE
MLBr_00315|M.leprae_Br4923 ------------LFLTQDAATGALPTLRAATDAAVL--GGQYFGPDGFAE
Mb2286|M.bovis_AF2122/97 RLY--KTSWRFAPFLWQEIEEGILPALYAAATPQAD--GGAFYGPRGRYE
Rv2263|M.tuberculosis_H37Rv RLY--KTSWRFAPFLWQEIEEGILPALYAAATPQAD--GGAFYGPRGRYE
MMAR_3355|M.marinum_M RFY--TTSWRFAPFLWQEIEDGILPALYAAVTPQAE--GGAFYGPRGFYE
MUL_1288|M.ulcerans_Agy99 RFY--TTSWRFAPFLWQEIEDGILPALYAAVTPQAE--GGAFYGPRGFYE
Mvan_0945|M.vanbaalenii_PYR-1 DG---GQPIVDPDRDMKTPQQGASTQVFAATSPQLADIGGVYLANNDIAP
Mflv_2120|M.gilvum_PYR-GCK ASRNPDKPTTDFRKQFTTPEHGAATQVWAAVSDDLDGQGGLYLSDCRIR-
* . : **. *. : .
MAV_4710|M.avium_104 Q------------RGR-PKLVESSAQSHDEQLQRRLWAVSEELTGVHFPV
TH_4179|M.thermoresistible__bu I------------RGH-PVLVGSSAKSRDEDIQRRLWTVSEELTGVTFPV
MAB_0646c|M.abscessus_ATCC_199 Q------------RGN-PKVVASSEQSYNLDLQRRLWSVSEELTDVVFPV
MSMEG_0876|M.smegmatis_MC2_155 T------------RGY-PKVVASSAQSHDADLQRRLWAVSEELTGVTFPV
MLBr_00315|M.leprae_Br4923 I------------RGH-PKVVASNGKSHDVDRQLRLWAVSEELTGVVYPV
Mb2286|M.bovis_AF2122/97 V------------AGGGVREAKVPAAARNDADSKRLWEVSEQLTGVSYPK
Rv2263|M.tuberculosis_H37Rv V------------AGGGVREAKVPAAARNDADSKRLWEVSEQLTGVSYPK
MMAR_3355|M.marinum_M A------------AGGGVRAAKVPARAGNDADCQRLWEVSERLTGVGYPT
MUL_1288|M.ulcerans_Agy99 A------------AGGGVRAAKVPARAGNDADCQRLWEVSERLTGVGYPT
Mvan_0945|M.vanbaalenii_PYR-1 LEADATPVTVDMGAGPLQTTPGVTGYAVDPESAERLWELSEKLLA-----
Mflv_2120|M.gilvum_PYR-GCK -----------------EAAP----YAADETRALTLWALSEHLCTASSAT
: : ** :**.*
MAV_4710|M.avium_104 -------
TH_4179|M.thermoresistible__bu -------
MAB_0646c|M.abscessus_ATCC_199 K------
MSMEG_0876|M.smegmatis_MC2_155 GEALQQA
MLBr_00315|M.leprae_Br4923 G------
Mb2286|M.bovis_AF2122/97 SR-----
Rv2263|M.tuberculosis_H37Rv SR-----
MMAR_3355|M.marinum_M SS-----
MUL_1288|M.ulcerans_Agy99 SS-----
Mvan_0945|M.vanbaalenii_PYR-1 -------
Mflv_2120|M.gilvum_PYR-GCK -------