For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTTSETTALDIVAGADLGGKTCVITGASSGLGRESARALASTGAHVILAARNLEALSDTEAWVRAEVADA ELSIVPLDLTSLASVASAAAQISELTPAVHVLMNNAGVMFTPFGRTAEGFETQFGTNHLGHFEFTRLLFP ALVAADGARVVNLSSEGHRISDVDFEDPNWETRDYDKFAAYGASKTANVLHAVELDRRLRDSGVRAFAVH PGIVATSLARHMTNDDFASLNKSTASRNPDKPTTDFRKQFTTPEHGAATQVWAAVSDDLDGQGGLYLSDC RIREAAPYAADETRALTLWALSEHLCTASSAT*
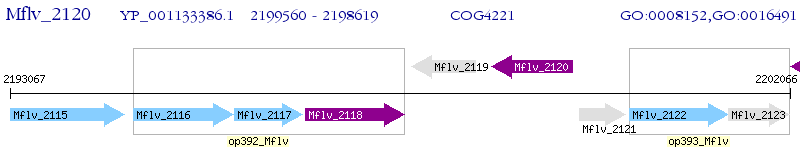
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2120 | - | - | 100% (313) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_0143 | - | 3e-44 | 46.41% (209) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0151 | - | 1e-42 | 42.06% (233) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0069 | - | 5e-50 | 51.38% (218) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0068 | - | 1e-49 | 50.92% (218) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 4e-45 | 39.74% (302) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0646c | - | 6e-48 | 46.61% (221) | short chain dehydrogenase |
| M. marinum M | MMAR_0757 | - | 6e-45 | 47.87% (211) | dehydrogenase/reductase |
| M. avium 104 | MAV_4710 | - | 2e-46 | 40.92% (303) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0876 | - | 4e-49 | 48.56% (208) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4179 | - | 7e-44 | 37.01% (308) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1391 | - | 3e-45 | 47.87% (211) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4584 | - | 1e-148 | 82.96% (311) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0757|M.marinum_M ----------------MSANGKAKSHWSASDIPD---QSGRVVVVTGANT
MUL_1391|M.ulcerans_Agy99 ----------------MSAKGKAKSHWSASDIPD---QSGRVVVVTGANT
MAV_4710|M.avium_104 ----------------MAPHDKRHRDWSEADVGD---QSGRVVVITGANT
Mb0069|M.bovis_AF2122/97 -----------------------MTKWTAADIPD---QTGRTAVITGANT
Rv0068|M.tuberculosis_H37Rv -----------------------MTKWTAADIPD---QTGRTAVITGANT
MAB_0646c|M.abscessus_ATCC_199 -------------------MSRSTTKWSTTDIPD---QTGRIAIVTGANT
MSMEG_0876|M.smegmatis_MC2_155 -----------------------MTKWTTARIPD---QTGRVAIVTGANT
MLBr_00315|M.leprae_Br4923 -----------------------MAKWTTADIPD---QTGRVAVITGANT
TH_4179|M.thermoresistible__bu ----------------------MTGKWTARDVPD---QTGRTAVITGANT
Mflv_2120|M.gilvum_PYR-GCK --------------------MTTTSETTALDIVAGADLGGKTCVITGASS
Mvan_4584|M.vanbaalenii_PYR-1 MGVIVGPGEHDPAAACDPHVVTATSAPTALDIVEGVDLSGKTCVITGASS
: : *: ::***.:
MMAR_0757|M.marinum_M GLGYHTAEALAGRGAHVVLAVRNPEKGNAAVAQIVAAKPQADVTLQALDL
MUL_1391|M.ulcerans_Agy99 GLGYHTAEALADRGAHVVLAVRNPEKGNAAVAQIVAAKPQADVTLQALDL
MAV_4710|M.avium_104 GIGYETAAVLAHRGAHVVLAVRDLEKGNAALSRIVAASPNADVTLQQLDL
Mb0069|M.bovis_AF2122/97 GLGFETAAALAAHGAHVVLAVRNLDKGKQAAARITEATPGAEVELQELDL
Rv0068|M.tuberculosis_H37Rv GLGFETAAALAAHGAHVVLAVRNLDKGKQAAARITEATPGAEVELQELDL
MAB_0646c|M.abscessus_ATCC_199 GLGLETAKALAAHGAHVVLAVRNAEKGKAAAEAITAAHSNADVTLQSLDL
MSMEG_0876|M.smegmatis_MC2_155 GLGLETAKALAAKGAHVVLAVRNLDKGKAAVDWIARSAPTADLELQQLDL
MLBr_00315|M.leprae_Br4923 GLGYQTALALAEHGAHVVLAVRNLDKGKDAAARITATSAQNNVALQELDL
TH_4179|M.thermoresistible__bu GLGFETAKVLAEKGAHVVLAVRDPDKGRRAADRITAAAPHADVTVRQLDL
Mflv_2120|M.gilvum_PYR-GCK GLGRESARALASTGAHVILAARNLEALSDTEAWVRAEVADAELSIVPLDL
Mvan_4584|M.vanbaalenii_PYR-1 GLGRESARALAKAGAHVILAARNAAAMSETEAWLRAQLPDALLSVVDLDL
*:* .:* .** ****:**.*: : : . : : ***
MMAR_0757|M.marinum_M SSLDSVRSAADALRSAYPRIDLLINNAGVMWTPKQVTKDGFEMQFGTNHL
MUL_1391|M.ulcerans_Agy99 SSLDSVRSAADALRSAYPRIDLLINNAGVMWTPKQVTKDGFEMQFGTNHL
MAV_4710|M.avium_104 ASLASVRSAAEALRAAYPRIDLLINNAGVMWTPKQVTEDGFELQFGTNHL
Mb0069|M.bovis_AF2122/97 TSLASVRAAAAQLKSDHQRIDLLINNAGVMYTPRQTTADGFEMQFGTNHL
Rv0068|M.tuberculosis_H37Rv TSLASVRAAAAQLKSDHQRIDLLINNAGVMYTPRQTTADGFEMQFGTNHL
MAB_0646c|M.abscessus_ATCC_199 SSLESVRRASDELKGRYDKIDLLINNAGVMWTEKSSTADGFELQFGTNHL
MSMEG_0876|M.smegmatis_MC2_155 GSLASVRAAADDLKGKFDRIDLLVNNAGVMWPPRQTTADGFELQFGTNHL
MLBr_00315|M.leprae_Br4923 ASLESVRAAAKQLRSDYDHIDLLINNAGVMWTPKSTTKDGFELQFGTNHL
TH_4179|M.thermoresistible__bu TSLDNIRRAADDLRAGYPRIDLLINNAGVMYPPRQTTRDGFELQFGTNHL
Mflv_2120|M.gilvum_PYR-GCK TSLASVASAAAQISELTPAVHVLMNNAGVMFTPFGRTAEGFETQFGTNHL
Mvan_4584|M.vanbaalenii_PYR-1 TSLASIAAAAAEISELTPVVHVLMNNAGVMFTPFGRTVDGFEIQFGTNHL
** .: *: : :.:*:******:. * :*** *******
MMAR_0757|M.marinum_M GHFALTGLLLDHLLPVPGSRVITVSSLGHRIRAAIHFDDLQWER-SYNRV
MUL_1391|M.ulcerans_Agy99 GHFALTGLLLDHLLPVPGSRVITVSSLGHRIRAAIHFDDLQWER-SYNRV
MAV_4710|M.avium_104 GHFALTGLLLDHLLGVRDSRVVTVSSLGHRLRAAIHFDDLHWER-RYDRV
Mb0069|M.bovis_AF2122/97 GHFALTGLLIDRLLPVAGSRVVTISSVGHRIRAAIHFDDLQWER-RYRRV
Rv0068|M.tuberculosis_H37Rv GHFALTGLLIDRLLPVAGSRVVTISSVGHRIRAAIHFDDLQWER-RYRRV
MAB_0646c|M.abscessus_ATCC_199 GHYALTGLLLERLLPVEGSRVVTVSSIGHRIRADIHFDDLQWER-DYDRV
MSMEG_0876|M.smegmatis_MC2_155 GHFALTGLLLDRMLTVPGSRVVTVSSQGHRILAAIHFDDLQWER-RYNRV
MLBr_00315|M.leprae_Br4923 GHFAFTGLLLDRLLPIVGSRVITVSSLSHRLFADIHFNDLQWEC-NYNRV
TH_4179|M.thermoresistible__bu GHFALTGQLLDNILPVDGSRVVTVASIAHRNMADIHFDDLQWER-GYHRV
Mflv_2120|M.gilvum_PYR-GCK GHFEFTRLLFPALVAADGARVVNLSSEGHRIS-DVDFEDPNWETRDYDKF
Mvan_4584|M.vanbaalenii_PYR-1 GHFELTRLLFPALVAADGARVVNLSSEGHRMG-DVDFDDPNWEHRDYDKF
**: :* *: :: .:**:.::* .** :.*:* :** * :.
MMAR_0757|M.marinum_M AAYGQSKLANLLFTYELQRRLAADSQAATIAVAAHPGGSNTELARNLPRM
MUL_1391|M.ulcerans_Agy99 AAYGQSKLANLLFTYELQRRLAADSQAATIAVAAHPGDSNTELARNLPRM
MAV_4710|M.avium_104 AAYGQSKLANLLFTYELQRRLAAAPDAKTIAVAAHPGGSNTELARHLPGI
Mb0069|M.bovis_AF2122/97 AAYGQAKLANLLFTYELQRRLAP--GGTTIAVASHPGVSNTELVRNMPRP
Rv0068|M.tuberculosis_H37Rv AAYGQAKLANLLFTYELQRRLAP--GGTTIAVASHPGVSNTEVVRNMPRP
MAB_0646c|M.abscessus_ATCC_199 AAYGQSKLANLLFTYELQRRLA---GTNTVALAAHPGGSNTELARNSPLW
MSMEG_0876|M.smegmatis_MC2_155 AAYGQSKLANLLFTYELQRRLT---GHQTTALAAHPGASNTELARHLP--
MLBr_00315|M.leprae_Br4923 AAYGQSKLANLLFTYELQRRLAT--RQTTIAVAAHPGGSRTELTRTLPAL
TH_4179|M.thermoresistible__bu AAYGQSKLANLMFAYELQRRLSAK-NAPTISVAAHPGVSNTELTRYIPGA
Mflv_2120|M.gilvum_PYR-GCK AAYGASKTANVLHAVELDRRLR---DSGVRAFAVHPGIVATSLARHMTND
Mvan_4584|M.vanbaalenii_PYR-1 AAYGASKTANVLHAVELDRRLR---DSGVRAFAVHPGIVATSLARHMTND
**** :* **::.: **:*** . :.* *** *.:.* .
MMAR_0757|M.marinum_M LVPLANILGPALFQSAQMGALPTLRAATDPSVAGGQYYGPDGFAEQRG--
MUL_1391|M.ulcerans_Agy99 LVPLANILGPALFQSAQMGALPTLRTATDPSAAGGQYYGPDGFAEQRG--
MAV_4710|M.avium_104 FRPVQAVLGPVLFQSPAMGALPTLRAATDPAVQGAQYYGPDGFLEQRG--
Mb0069|M.bovis_AF2122/97 LVAVAAILAP-LMQDAELGALPTLRAATDPAVRGGQYFGPDGFGEIRG--
Rv0068|M.tuberculosis_H37Rv LVAVAAILAP-LMQDAELGALPTLRAATDPAVRGGQYFGPDGFGEIRG--
MAB_0646c|M.abscessus_ATCC_199 VRAVFDVVAPVLVQGADMGALPTLRAATDPAALGGQYYGPDGFMEQRG--
MSMEG_0876|M.smegmatis_MC2_155 --GALERLVTPLAQDAALGALPTLRAATDPGALGGQYFGPDGIGETRG--
MLBr_00315|M.leprae_Br4923 IAPIFSVAELFLTQDAATGALPTLRAATDAAVLGGQYFGPDGFAEIRG--
TH_4179|M.thermoresistible__bu RLPGVSLLAGLLTNSPAVGALATLRAATDPEVKGGQYYGPDGFQEIRG--
Mflv_2120|M.gilvum_PYR-GCK DFASLNKSTASRNPDKPTTDFRKQFTTPEHGAATQVWAAVSDDLDGQGGL
Mvan_4584|M.vanbaalenii_PYR-1 DFASLNAASSTRNSDKPATDFRKQFTTPEYGAATQVWAAVSTDLDDAGGV
. : . ::.: . : . . : *
MMAR_0757|M.marinum_M ---HPKIVQSSAQSHDEDLQRRLWTVSEELTGVSFPV-------
MUL_1391|M.ulcerans_Agy99 ---HPKIVQSSAQSHDEDLQRRLWTVSEELTGVSFPV-------
MAV_4710|M.avium_104 ---RPKLVESSAQSHDEQLQRRLWAVSEELTGVHFPV-------
Mb0069|M.bovis_AF2122/97 ---YPKVVASSAQSHDEQLQRRLWAVSEELTGVVYPVG------
Rv0068|M.tuberculosis_H37Rv ---YPKVVASSAQSHDEQLQRRLWAVSEELTGVVYPVG------
MAB_0646c|M.abscessus_ATCC_199 ---NPKVVASSEQSYNLDLQRRLWSVSEELTDVVFPVK------
MSMEG_0876|M.smegmatis_MC2_155 ---YPKVVASSAQSHDADLQRRLWAVSEELTGVTFPVGEALQQA
MLBr_00315|M.leprae_Br4923 ---HPKVVASNGKSHDVDRQLRLWAVSEELTGVVYPVG------
TH_4179|M.thermoresistible__bu ---HPVLVGSSAKSRDEDIQRRLWTVSEELTGVTFPV-------
Mflv_2120|M.gilvum_PYR-GCK YLSDCRIREAAPYAADETRALTLWALSEHLCTASSAT-------
Mvan_4584|M.vanbaalenii_PYR-1 YLSDCEVRRAAAYAVDEARALALWDLSERLCTRSSPSLGSGA--
: : : : ** :**.* .